6O3A
 
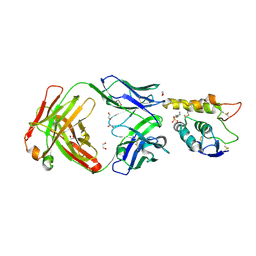 | | Crystal structure of Frizzled 7 CRD in complex with F7.B Fab | | 分子名称: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, ... | | 著者 | Raman, S, Beilschmidt, M, Fransson, J, Julien, J.P. | | 登録日 | 2019-02-26 | | 公開日 | 2019-04-03 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structure-guided design fine-tunes pharmacokinetics, tolerability, and antitumor profile of multispecific frizzled antibodies.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
6HZU
 
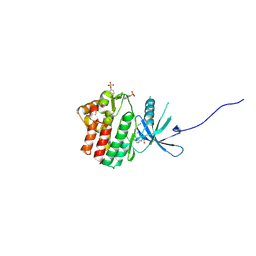 | | HUMAN JAK1 IN COMPLEX WITH LASW1393 | | 分子名称: | 2-[4-[8-oxidanylidene-2-[(~{E})-(2-oxidanylidenepyridin-3-ylidene)amino]-7~{H}-purin-9-yl]cyclohexyl]ethanenitrile, Tyrosine-protein kinase JAK1 | | 著者 | Lozoya, E, Segarra, V, Bach, J, Jestel, A, Lammens, A, Blaesse, M. | | 登録日 | 2018-10-24 | | 公開日 | 2019-10-23 | | 最終更新日 | 2019-11-06 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Identification of 2-Imidazopyridine and 2-Aminopyridone Purinones as Potent Pan-Janus Kinase (JAK) Inhibitors for the Inhaled Treatment of Respiratory Diseases.
J.Med.Chem., 62, 2019
|
|
4R1T
 
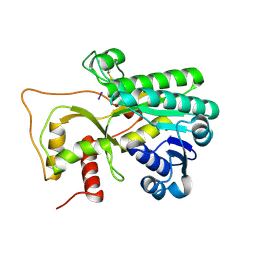 | | Crystal structure of Petunia hydrida cinnamoyl-CoA reductase | | 分子名称: | cinnamoyl CoA reductase, molecular iodine | | 著者 | Noel, J.P, Louie, G.V, Bowman, M.E, Bomati, E.K. | | 登録日 | 2014-08-07 | | 公開日 | 2014-10-01 | | 最終更新日 | 2014-11-12 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structural Studies of Cinnamoyl-CoA Reductase and Cinnamyl-Alcohol Dehydrogenase, Key Enzymes of Monolignol Biosynthesis.
Plant Cell, 26, 2014
|
|
4R1U
 
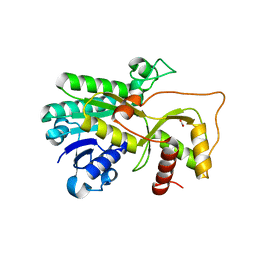 | | Crystal structure of Medicago truncatula cinnamoyl-CoA reductase | | 分子名称: | ACETATE ION, Cinnamoyl CoA reductase | | 著者 | Noel, J.P, Bomati, E.K, Louie, G.V, Bowman, M.E. | | 登録日 | 2014-08-07 | | 公開日 | 2014-10-01 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.18 Å) | | 主引用文献 | Structural Studies of Cinnamoyl-CoA Reductase and Cinnamyl-Alcohol Dehydrogenase, Key Enzymes of Monolignol Biosynthesis.
Plant Cell, 26, 2014
|
|
4R1S
 
 | | Crystal structure of Petunia hydrida cinnamoyl-CoA reductase | | 分子名称: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, cinnamoyl CoA reductase | | 著者 | Noel, J.P, Louie, G.V, Bowman, M.E, Bomati, E.K. | | 登録日 | 2014-08-07 | | 公開日 | 2014-10-01 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Structural Studies of Cinnamoyl-CoA Reductase and Cinnamyl-Alcohol Dehydrogenase, Key Enzymes of Monolignol Biosynthesis.
Plant Cell, 26, 2014
|
|
7K0V
 
 | | Crystal structure of bRaf in complex with inhibitor GNE-0749 | | 分子名称: | CHLORIDE ION, N-(3,3-dimethylbutyl)-N'-{2-fluoro-5-[(5-fluoro-3-methyl-4-oxo-3,4-dihydroquinazolin-6-yl)amino]-4-methylphenyl}urea, Non-specific serine/threonine protein kinase | | 著者 | Yin, J, Eigenbrot, C.E, Wang, W. | | 登録日 | 2020-09-06 | | 公開日 | 2021-05-26 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.93 Å) | | 主引用文献 | Targeting KRAS Mutant Cancers via Combination Treatment: Discovery of a 5-Fluoro-4-(3 H )-quinazolinone Aryl Urea pan-RAF Kinase Inhibitor.
J.Med.Chem., 64, 2021
|
|
7X0E
 
 | | Structure of Pseudomonas NRPS protein, AmbB-TC in apo form | | 分子名称: | AMB antimetabolite synthase AmbB, N-methyl-N-[(2S,3R,4R,5R)-2,3,4,5,6-pentakis(oxidanyl)hexyl]nonanamide | | 著者 | ChuYuanKee, M, Bharath, S.R, Song, H. | | 登録日 | 2022-02-22 | | 公開日 | 2022-07-06 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structural insights into the substrate-bound condensation domains of non-ribosomal peptide synthetase AmbB.
Sci Rep, 12, 2022
|
|
6XF6
 
 | | Cryo-EM structure of a biotinylated SARS-CoV-2 spike probe in the prefusion state (1 RBD up) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | 著者 | Cerutti, G, Gorman, J, Kwong, P.D, Shapiro, L. | | 登録日 | 2020-06-15 | | 公開日 | 2020-09-02 | | 最終更新日 | 2020-12-02 | | 実験手法 | ELECTRON MICROSCOPY (4 Å) | | 主引用文献 | Structure-Based Design with Tag-Based Purification and In-Process Biotinylation Enable Streamlined Development of SARS-CoV-2 Spike Molecular Probes.
SSRN, 2020
|
|
7OBN
 
 | |
7SVI
 
 | |
4IWD
 
 | | Structure of dually phosphorylated c-MET receptor kinase in complex with an MK-8033 analog | | 分子名称: | 1-{5-oxo-3-[1-(piperidin-4-yl)-1H-pyrazol-4-yl]-5H-benzo[4,5]cyclohepta[1,2-b]pyridin-7-yl}-N-(pyridin-2-ylmethyl)methanesulfonamide, Hepatocyte growth factor receptor | | 著者 | Soisson, S.M, Northrup, A, Rickert, K, Patel, S, Allison, T. | | 登録日 | 2013-01-23 | | 公開日 | 2013-12-11 | | 最終更新日 | 2017-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.99 Å) | | 主引用文献 | Discovery of 1-[3-(1-methyl-1H-pyrazol-4-yl)-5-oxo-5H-benzo[4,5]cyclohepta[1,2-b]pyridin-7-yl]-N-(pyridin-2-ylmethyl)methanesulfonamide (MK-8033): A Specific c-Met/Ron dual kinase inhibitor with preferential affinity for the activated state of c-Met.
J.Med.Chem., 56, 2013
|
|
7JNR
 
 | | Carbonic Anhydrase II Complexed with N-(5-sulfamoyl-1,3,4-thiadiazol-2-yl)propionamide | | 分子名称: | Carbonic anhydrase 2, GLYCEROL, N-(5-sulfamoyl-1,3,4-thiadiazol-2-yl)propanamide, ... | | 著者 | Andring, J.T, McKenna, R. | | 登録日 | 2020-08-05 | | 公開日 | 2020-11-04 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.443 Å) | | 主引用文献 | Structural Basis of Nanomolar Inhibition of Tumor-Associated Carbonic Anhydrase IX: X-Ray Crystallographic and Inhibition Study of Lipophilic Inhibitors with Acetazolamide Backbone.
J.Med.Chem., 63, 2020
|
|
7JNW
 
 | | Carbonic Anhydrase II Complexed with N-(5-sulfamoyl-1,3,4-thiadiazol-2-yl)isobutyramide | | 分子名称: | 2-methyl-N-(5-sulfamoyl-1,3,4-thiadiazol-2-yl)propanamide, Carbonic anhydrase 2, DIMETHYL SULFOXIDE, ... | | 著者 | Andring, J.T, McKenna, R. | | 登録日 | 2020-08-05 | | 公開日 | 2020-11-04 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.292 Å) | | 主引用文献 | Structural Basis of Nanomolar Inhibition of Tumor-Associated Carbonic Anhydrase IX: X-Ray Crystallographic and Inhibition Study of Lipophilic Inhibitors with Acetazolamide Backbone.
J.Med.Chem., 63, 2020
|
|
1NBS
 
 | | Crystal structure of the specificity domain of Ribonuclease P RNA | | 分子名称: | LEAD (II) ION, MAGNESIUM ION, RIBONUCLEASE P RNA | | 著者 | Krasilnikov, A.S, Yang, X, Pan, T, Mondragon, A. | | 登録日 | 2002-12-03 | | 公開日 | 2003-02-18 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (3.15 Å) | | 主引用文献 | Crystal structure of the specificity domain of Ribonuclease P
Nature, 421, 2003
|
|
3QNQ
 
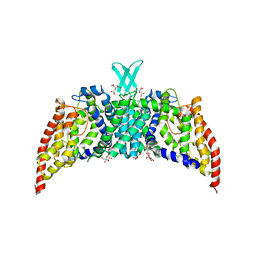 | | Crystal structure of the transporter ChbC, the IIC component from the N,N'-diacetylchitobiose-specific phosphotransferase system | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CITRIC ACID, PTS system, ... | | 著者 | Cao, Y, Jin, X, Huang, H, Levin, E.J, Zhou, M, New York Consortium on Membrane Protein Structure (NYCOMPS) | | 登録日 | 2011-02-08 | | 公開日 | 2011-04-06 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (3.295 Å) | | 主引用文献 | Crystal structure of a phosphorylation-coupled saccharide transporter.
Nature, 473, 2011
|
|
7SVH
 
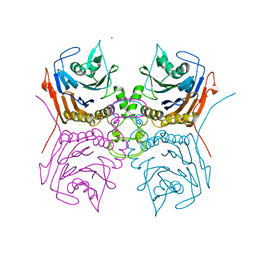 | |
7SVJ
 
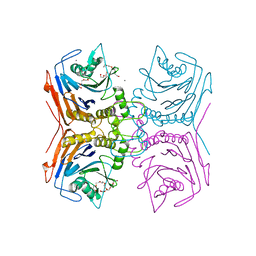 | | Bile Salt Hydrolase from Lactobacillus ingluviei | | 分子名称: | CALCIUM ION, Choloylglycine hydrolase, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Walker, M.E, Patel, S, Redinbo, M.R. | | 登録日 | 2021-11-19 | | 公開日 | 2023-01-25 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.43 Å) | | 主引用文献 | Bile salt hydrolases shape the bile acid landscape and restrict Clostridioides difficile growth in the murine gut.
Nat Microbiol, 8, 2023
|
|
7SVG
 
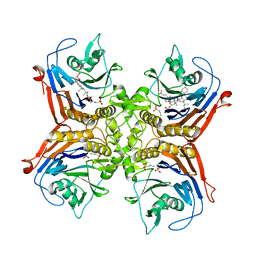 | |
7SVF
 
 | |
7SVE
 
 | |
1N7A
 
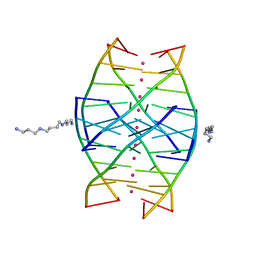 | | RIP-Radiation-damage Induced Phasing | | 分子名称: | POTASSIUM ION, RNA/DNA (5'-R(*U)-D(P*(BGM))-R(P*AP*GP*GP*U)-3'), SPERMINE | | 著者 | Ravelli, R.B.G, Leiros, H.-K.S, Pan, B, Caffrey, M, McSweeney, S. | | 登録日 | 2002-11-13 | | 公開日 | 2003-03-04 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.2 Å) | | 主引用文献 | Specific Radiation-Damage Can Be Used To Solve Macromolecular Crystal Structures
Structure, 11, 2003
|
|
1OSX
 
 | | Solution Structure of the Extracellular Domain of BLyS Receptor 3 (BR3) | | 分子名称: | Tumor necrosis factor receptor superfamily member 13C | | 著者 | Gordon, N.C, Pan, B, Hymowitz, S.G, Yin, J.P, Kelley, R.F, Cochran, A.G, Yan, M, Dixit, V.M, Fairbrother, W.J, Starovasnik, M.A. | | 登録日 | 2003-03-20 | | 公開日 | 2003-05-27 | | 最終更新日 | 2022-02-23 | | 実験手法 | SOLUTION NMR | | 主引用文献 | BAFF/BLyS receptor 3 comprises a minimal TNF receptor-like module that encodes a highly focused ligand-binding site
Biochemistry, 42, 2003
|
|
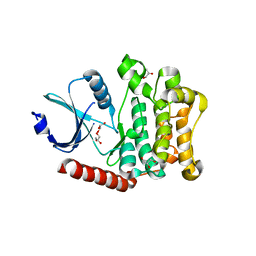
4LG4
 
 | |
1OSG
 
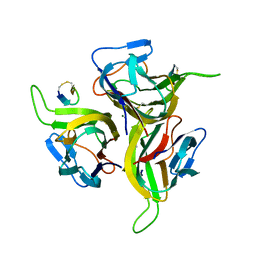 | | Complex between BAFF and a BR3 derived peptide presented in a beta-hairpin scaffold | | 分子名称: | BR3 derived PEPTIDE, MAGNESIUM ION, Tumor necrosis factor ligand superfamily member 13B | | 著者 | Gordon, N.C, Pan, B, Hymowitz, S.G, Yin, J.P, Kelley, R.F, Cochran, A.G, Yan, M, Dixit, V.M, Fairbrother, W.J, Starovasnik, M.A. | | 登録日 | 2003-03-19 | | 公開日 | 2003-05-27 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | BAFF/BLyS receptor 3 comprises a minimal TNF receptor-like module that encodes a highly focused ligand-binding site
Biochemistry, 42, 2003
|
|
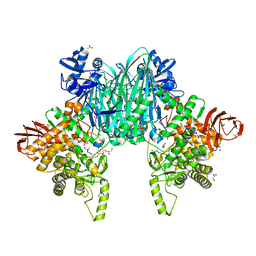
8H2W
 
 | |
