5V8T
 
 | | Crystal structure of SMT fusion Peptidyl-prolyl cis-trans isomerase from Burkholderia pseudomallei complexed with SF354 | | 分子名称: | 2-{[3,5-bis(2-methoxyethoxy)benzene-1-carbonyl]amino}ethyl (2S)-1-(benzylsulfonyl)piperidine-2-carboxylate, FORMIC ACID, MAGNESIUM ION, ... | | 著者 | Lorimer, D.D, Dranow, D.M, Seufert, F, Abendroth, J, Holzgrabe, U. | | 登録日 | 2017-03-22 | | 公開日 | 2018-03-14 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Crystal structure of SMT fusion Peptidyl-prolyl cis-trans isomerase from Burkholderia pseudomallei complexed with SF354
to be published
|
|
5OM9
 
 | | Crystal structure of the human CARBOXYPEPTIDASE A1 in complex with a thiirane mechanism-based inhibitor | | 分子名称: | (2~{R})-4-methyl-2-[(1~{S})-1-sulfanylethyl]pentanoic acid, Carboxypeptidase A1, ZINC ION | | 著者 | Gallego, P, Granados, C, Fernandez, D, Pallares, I, Covaleda, G, Aviles, F.X, Vendrell, J, Reverter, D. | | 登録日 | 2017-07-28 | | 公開日 | 2017-08-09 | | 最終更新日 | 2025-10-01 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Discovery of Mechanism-Based Inactivators for Human Pancreatic Carboxypeptidase A from a Focused Synthetic Library.
ACS Med Chem Lett, 8, 2017
|
|
3RCL
 
 | | Human Cyclophilin D Complexed with a Fragment | | 分子名称: | 3-(1,3-oxazol-5-yl)aniline, O-ACETALDEHYDYL-HEXAETHYLENE GLYCOL, Peptidyl-prolyl cis-trans isomerase F, ... | | 著者 | Colliandre, L, Ahmed-Belkacem, H, Bessin, Y, Pawlotsky, J.M, Guichou, J.F. | | 登録日 | 2011-03-31 | | 公開日 | 2012-03-21 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Fragment-based discovery of a new family of non-peptidic small-molecule cyclophilin inhibitors with potent antiviral activities.
Nat Commun, 7, 2016
|
|
1N8N
 
 | | Crystal structure of the Au3+ complex of AphA class B acid phosphatase/phosphotransferase from E. coli at 1.69 A resolution | | 分子名称: | Class B acid phosphatase, GOLD 3+ ION | | 著者 | Calderone, V, Forleo, C, Benvenuti, M, Rossolini, G.M, Thaller, M.C, Mangani, S. | | 登録日 | 2002-11-21 | | 公開日 | 2004-02-03 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.69 Å) | | 主引用文献 | The first structure of a bacterial class B Acid phosphatase reveals further structural heterogeneity among phosphatases of the haloacid dehalogenase fold.
J.Mol.Biol., 335, 2004
|
|
6B4D
 
 | | Crystal structure of human carbonic anhydrase II in complex with a heteroaryl-pyrazole carboxylic acid derivative. | | 分子名称: | 3-(1-ethyl-1H-indol-3-yl)-1-methyl-1H-pyrazole-5-carboxylic acid, Carbonic anhydrase 2, DIMETHYL SULFOXIDE, ... | | 著者 | Lomelino, C.L, Mahon, B.P, McKenna, R. | | 登録日 | 2017-09-26 | | 公開日 | 2018-02-07 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.196 Å) | | 主引用文献 | Exploring Heteroaryl-pyrazole Carboxylic Acids as Human Carbonic Anhydrase XII Inhibitors.
ACS Med Chem Lett, 8, 2017
|
|
4MXV
 
 | | Structure of Lymphotoxin alpha bound to anti-LTa Fab | | 分子名称: | Lymphotoxin-alpha, anti-Lymphotoxin alpha antibody heavy chain, anti-Lymphotoxin alpha antibody light chain | | 著者 | Yin, J.P, Hymowitz, S.G. | | 登録日 | 2013-09-26 | | 公開日 | 2013-11-13 | | 最終更新日 | 2024-10-09 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Dimerization of LT beta R by LT alpha 1 beta 2 is necessary and sufficient for signal transduction.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3MSK
 
 | | Fragment Based Discovery and Optimisation of BACE-1 Inhibitors | | 分子名称: | 4-(2-amino-5-chloro-1H-benzimidazol-1-yl)-N-cyclohexyl-N-methylbutanamide, Beta-secretase 1, GLYCEROL, ... | | 著者 | Smith, M.A, Madden, J.M, Barker, J, Godemann, R, Kraemer, J, Hallett, D. | | 登録日 | 2010-04-29 | | 公開日 | 2010-07-14 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Fragment-based discovery and optimization of BACE1 inhibitors.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
6B1W
 
 | | Crystal structure KPC-2 beta-lactamase complexed with WCK 5107 by co-crystallization | | 分子名称: | (2S,5R)-1-formyl-N'-[(3R)-piperidine-3-carbonyl]-5-[(sulfooxy)amino]piperidine-2-carbohydrazide, CHLORIDE ION, Carbapenem-hydrolyzing beta-lactamase KPC, ... | | 著者 | van den Akker, F, Nguyen, N.Q. | | 登録日 | 2017-09-19 | | 公開日 | 2018-08-01 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (1.73 Å) | | 主引用文献 | Strategic Approaches to Overcome Resistance against Gram-Negative Pathogens Using beta-Lactamase Inhibitors and beta-Lactam Enhancers: Activity of Three Novel Diazabicyclooctanes WCK 5153, Zidebactam (WCK 5107), and WCK 4234.
J. Med. Chem., 61, 2018
|
|
1O4B
 
 | | CRYSTAL STRUCTURE OF SH2 IN COMPLEX WITH RU83876. | | 分子名称: | N-ACETYL-N-[1-(1,1'-BIPHENYL-4-YLMETHYL)-2-OXOAZEPAN-3-YL]-3,4-DIPHOSPHONOPHENYLALANINAMIDE, PROTO-ONCOGENE TYROSINE-PROTEIN KINASE SRC | | 著者 | Lange, G, Loenze, P, Liesum, A. | | 登録日 | 2003-06-15 | | 公開日 | 2004-02-17 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Requirements for specific binding of low affinity inhibitor fragments to the SH2 domain of (pp60)Src are identical to those for high affinity binding of full length inhibitors.
J.Med.Chem., 46, 2003
|
|
5VBS
 
 | | Structural basis for a six letter alphabet including GATCKX | | 分子名称: | DNA (5'-D(*CP*TP*TP*AP*TP*(DX)P*(DX)P*T)-3'), DNA (5'-D(P*AP*(93D)P*(93D)P*AP*TP*AP*AP*G)-3'), reverse transcriptase catalytic fragment | | 著者 | Singh, I, Georgiadis, M.M. | | 登録日 | 2017-03-30 | | 公開日 | 2018-03-07 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.749 Å) | | 主引用文献 | Structure and Biophysics for a Six Letter DNA Alphabet that Includes Imidazo[1,2-a]-1,3,5-triazine-2(8H)-4(3H)-dione (X) and 2,4-Diaminopyrimidine (K).
ACS Synth Biol, 6, 2017
|
|
3RIK
 
 | | The acid beta-glucosidase active site exhibits plasticity in binding 3,4,5,6-tetrahydroxyazepane-based inhibitors: implications for pharmacological chaperone design for gaucher disease | | 分子名称: | (3S,4R,5R,6S)-1-(2-hydroxyethyl)azepane-3,4,5,6-tetrol, 2-acetamido-2-deoxy-beta-D-glucopyranose, Glucosylceramidase, ... | | 著者 | Orwig, S.D, Lieberman, R.L. | | 登録日 | 2011-04-13 | | 公開日 | 2012-03-14 | | 最終更新日 | 2024-10-09 | | 実験手法 | X-RAY DIFFRACTION (2.48 Å) | | 主引用文献 | Binding of 3,4,5,6-tetrahydroxyazepanes to the acid-beta-glucosidase active site: implications for pharmacological chaperone design for Gaucher disease
Biochemistry, 50, 2011
|
|
6B63
 
 | | IMPase (AF2372) with 25 mM Asp | | 分子名称: | (2R)-2,3-dihydroxypropyl (2S)-2,3-dihydroxypropyl hydrogen phosphate, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Fructose-1,6-bisphosphatase/inositol-1-monophosphatase, ... | | 著者 | Goldstein, R.I, Roberts, M. | | 登録日 | 2017-10-01 | | 公開日 | 2018-10-03 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.696 Å) | | 主引用文献 | Osmolyte binding capacity of a dual action IMPase/FBPase (AF2372)
To Be Published
|
|
5URA
 
 | | Enantiomer-Specific Binding of the Potent Antinociceptive Agent SBFI-26 to Anandamide transporters FABP7 | | 分子名称: | (1S,2S,3S,4S)-3-{[(naphthalen-1-yl)oxy]carbonyl}-2,4-diphenylcyclobutane-1-carboxylic acid, Fatty acid-binding protein, brain, ... | | 著者 | Hsu, H.-C, Li, H. | | 登録日 | 2017-02-09 | | 公開日 | 2017-08-23 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.85002172 Å) | | 主引用文献 | The Antinociceptive Agent SBFI-26 Binds to Anandamide Transporters FABP5 and FABP7 at Two Different Sites.
Biochemistry, 56, 2017
|
|
1VKP
 
 | | X-RAY STRUCTURE OF GENE PRODUCT FROM ARABIDOPSIS THALIANA AT5G08170, AGMATINE IMINOHYDROLASE | | 分子名称: | 1,2-ETHANEDIOL, 3[N-MORPHOLINO]PROPANE SULFONIC ACID, AGMATINE IMINOHYDROLASE, ... | | 著者 | Wesenberg, G.E, Smith, D.W, Phillips Jr, G.N, Bingman, C.A, Allard, S.T.M, Center for Eukaryotic Structural Genomics (CESG) | | 登録日 | 2004-06-15 | | 公開日 | 2004-08-17 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (1.53 Å) | | 主引用文献 | X-ray Structure of Gene Product from Arabidopsis Thaliana At5g08170
To be published
|
|
2ZUM
 
 | |
4H77
 
 | | Crystal structure of haloalkane dehalogenase LinB from Sphingobium sp. MI1205 | | 分子名称: | CALCIUM ION, CHLORIDE ION, GLYCEROL, ... | | 著者 | Okai, M, Ohtsuka, J, Imai, F.L, Mase, T, Moriuchi, R, Tsuda, M, Nagata, K, Nagata, Y, Tanokura, M. | | 登録日 | 2012-09-20 | | 公開日 | 2013-06-05 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Crystal Structure and Site-Directed Mutagenesis Analyses of Haloalkane Dehalogenase LinB from Sphingobium sp. Strain MI1205.
J.Bacteriol., 195, 2013
|
|
167L
 
 | |
5SNL
 
 | | PanDDA analysis group deposition -- Crystal Structure of Pseudomonas Aeruginosa FabF-C164Q mutant protein in complex with Z2027049478 | | 分子名称: | 3-oxoacyl-[acyl-carrier-protein] synthase 2, 5-(1,3-thiazol-2-yl)-1H-1,2,4-triazole, DIMETHYL SULFOXIDE, ... | | 著者 | Brenk, R, Georgiou, C. | | 登録日 | 2022-05-30 | | 公開日 | 2023-12-20 | | 最終更新日 | 2025-05-07 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Towards new antibiotics: P. aeruginosa FabF ligands discovered by crystallographic fragment screening followed by hit expansion.
Eur.J.Med.Chem., 291, 2025
|
|
1RJE
 
 | | Structure of PPM1, a leucine carboxy methyltransferase involved in the regulation of protein phosphatase 2A activity | | 分子名称: | BETA-MERCAPTOETHANOL, S-ADENOSYL-L-HOMOCYSTEINE, SULFATE ION, ... | | 著者 | Leulliot, N, Quevillon-Cheruel, S, Sorel, I, de La Sierra-Gallay, I.L, Collinet, B, Graille, M, Blondeau, K, Bettache, N, Poupon, A, Janin, J, van Tilbeurgh, H. | | 登録日 | 2003-11-19 | | 公開日 | 2003-12-02 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structure of protein phosphatase methyltransferase 1 (PPM1), a leucine carboxyl methyltransferase involved in the regulation of protein phosphatase 2A activity.
J.Biol.Chem., 279, 2004
|
|
7GSQ
 
 | |
7GTD
 
 | |
7GSU
 
 | |
7GTR
 
 | |
8GC2
 
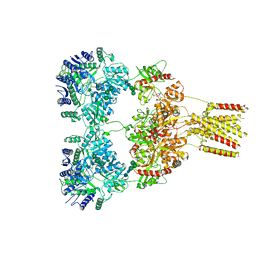 | | Domoate-bound GluK2 kainate receptor in partially-open conformation 1 | | 分子名称: | (2S,3S,4S)-2-CARBOXY-4-[(1Z,3E,5R)-5-CARBOXY-1-METHYL-1,3-HEXADIENYL]-3-PYRROLIDINEACETIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Bogdanovic, N, Tajima, N. | | 登録日 | 2023-03-01 | | 公開日 | 2024-03-13 | | 最終更新日 | 2024-10-16 | | 実験手法 | ELECTRON MICROSCOPY (4.1 Å) | | 主引用文献 | Structural basis for kainate receptor activation by a partial agonist
To Be Published
|
|
7GSY
 
 | |
