1PE4
 
 | | SOLUTION STRUCTURE OF TOXIN CN12 FROM CENTRUROIDES NOXIUS ALFA SCORPION TOXIN ACTING ON SODIUM CHANNELS. NMR STRUCTURE | | 分子名称: | Neurotoxin Cn11 | | 著者 | Del Rio-Portilla, F, Hernandez-Marin, E, Pimienta, G, Coronas, F.V, Zamudio, F.Z, Rodrguez de la Vega, R.C, Wanke, E, Possani, L.D. | | 登録日 | 2003-05-21 | | 公開日 | 2004-05-25 | | 最終更新日 | 2022-02-23 | | 実験手法 | SOLUTION NMR | | 主引用文献 | NMR solution structure of Cn12, a novel peptide from the Mexican scorpion Centruroides noxius with a typical beta-toxin sequence but with alpha-like physiological activity.
Eur.J.Biochem., 271, 2004
|
|
4NJC
 
 | |
1WFY
 
 | | Solution structure of the Ras-binding domain of mouse RGS14 | | 分子名称: | regulator of G-protein signaling 14; rap1/rap2 interacting protein | | 著者 | Nakanishi, T, Tochio, N, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2004-05-27 | | 公開日 | 2004-11-27 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of the Ras-binding domain of mouse RGS14
To be Published
|
|
1BMR
 
 | | ALPHA-LIKE TOXIN LQH III FROM SCORPION LEIURUS QUINQUESTRIATUS HEBRAEUS, NMR, 25 STRUCTURES | | 分子名称: | LQH III ALPHA-LIKE TOXIN | | 著者 | Krimm, I, Gilles, N, Sautiere, P, Stankiewicz, M, Pelhate, M, Gordon, D, Lancelin, J.-M. | | 登録日 | 1998-07-24 | | 公開日 | 1999-02-16 | | 最終更新日 | 2022-02-16 | | 実験手法 | SOLUTION NMR | | 主引用文献 | NMR structures and activity of a novel alpha-like toxin from the scorpion Leiurus quinquestriatus hebraeus.
J.Mol.Biol., 285, 1999
|
|
1CZ1
 
 | | EXO-B-(1,3)-GLUCANASE FROM CANDIDA ALBICANS AT 1.85 A RESOLUTION | | 分子名称: | PROTEIN (EXO-B-(1,3)-GLUCANASE) | | 著者 | Cutfield, S.M, Davies, G.J, Murshudov, G, Anderson, B.F, Moody, P.C.E, Sullivan, P.A, Cutfield, J.F. | | 登録日 | 1999-09-01 | | 公開日 | 2000-01-03 | | 最終更新日 | 2017-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | The structure of the exo-beta-(1,3)-glucanase from Candida albicans in native and bound forms: relationship between a pocket and groove in family 5 glycosyl hydrolases.
J.Mol.Biol., 294, 1999
|
|
4G33
 
 | | Crystal Structure of a Phospholipid-Lipoxygenase Complex from Pseudomonas aeruginosa at 2.0 A (C2221) | | 分子名称: | (2R)-3-{[(S)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-2-(tetradec-5-enoyloxy)propyl (11Z)-octadec-11-enoate, 15S-LIPOXYGENASE, FE (II) ION, ... | | 著者 | Carpena, X, Garreta, A, Val-Moraes, S.P, Garcia-Fernandez, Q, Fita, I. | | 登録日 | 2012-07-13 | | 公開日 | 2013-11-06 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.03 Å) | | 主引用文献 | Structure and interaction with phospholipids of a prokaryotic lipoxygenase from Pseudomonas aeruginosa.
Faseb J., 27, 2013
|
|
1C7T
 
 | | BETA-N-ACETYLHEXOSAMINIDASE MUTANT E540D COMPLEXED WITH DI-N ACETYL-D-GLUCOSAMINE (CHITOBIASE) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BETA-N-ACETYLHEXOSAMINIDASE, SULFATE ION | | 著者 | Prag, G, Papanikolau, Y, Tavlas, G, Vorgias, C.E, Petratos, K, Oppenheim, A.B. | | 登録日 | 2000-03-17 | | 公開日 | 2000-09-20 | | 最終更新日 | 2023-08-09 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structures of chitobiase mutants complexed with the substrate Di-N-acetyl-d-glucosamine: the catalytic role of the conserved acidic pair, aspartate 539 and glutamate 540.
J.Mol.Biol., 300, 2000
|
|
1C7S
 
 | | BETA-N-ACETYLHEXOSAMINIDASE MUTANT D539A COMPLEXED WITH DI-N-ACETYL-BETA-D-GLUCOSAMINE (CHITOBIASE) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BETA-N-ACETYLHEXOSAMINIDASE, SULFATE ION | | 著者 | Prag, G, Papanikolau, Y, Tavlas, G, Vorgias, C.E, Petratos, K, Oppenheim, A.B. | | 登録日 | 2000-03-14 | | 公開日 | 2000-09-20 | | 最終更新日 | 2023-08-09 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structures of chitobiase mutants complexed with the substrate Di-N-acetyl-d-glucosamine: the catalytic role of the conserved acidic pair, aspartate 539 and glutamate 540.
J.Mol.Biol., 300, 2000
|
|
1C7M
 
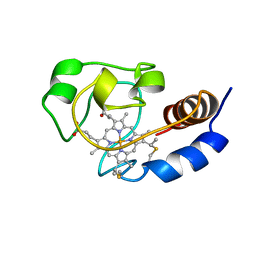 | | SOLUTION STRUCTURE OF THE FUNCTIONAL DOMAIN OF PARACOCCUS DENITRIFICANS CYTOCHROME C552 IN THE REDUCED STATE | | 分子名称: | HEME C, PROTEIN (CYTOCHROME C552) | | 著者 | Pristovsek, P, Luecke, C, Reincke, B, Ludwig, B, Rueterjans, H. | | 登録日 | 2000-03-03 | | 公開日 | 2000-07-19 | | 最終更新日 | 2023-12-27 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of the functional domain of Paracoccus denitrificans cytochrome c552 in the reduced state.
Eur.J.Biochem., 267, 2000
|
|
1WLZ
 
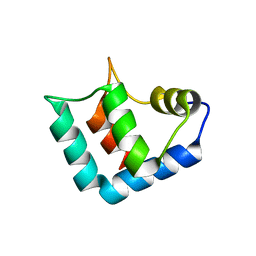 | | Crystal structure of DJBP fragment which was obtained by limited proteolysis | | 分子名称: | CAP-binding protein complex interacting protein 1 isoform a | | 著者 | Honbou, K, Suzuki, N, Horiuchi, M, Taira, T, Niki, T, Ariga, H, Inagaki, F. | | 登録日 | 2004-07-01 | | 公開日 | 2005-08-23 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Crystal Structure of DJBP Fragment which was obtained by Limited Proteolysis
To be Published
|
|
6RWG
 
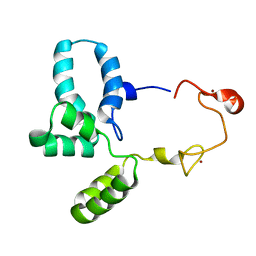 | | Structure of HIV-1 CAcSP1NC mutant(W41A,M42A) interacting with maturation inhibitor EP39 | | 分子名称: | Gag polyprotein, ZINC ION | | 著者 | Chen, X, Coric, P, Larue, V, Nonin-Lecomte, S, Bouaziz, S, Structural Genomics Consortium (SGC) | | 登録日 | 2019-06-05 | | 公開日 | 2020-05-20 | | 最終更新日 | 2024-06-19 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The HIV-1 maturation inhibitor, EP39, interferes with the dynamic helix-coil equilibrium of the CA-SP1 junction of Gag.
Eur.J.Med.Chem., 204, 2020
|
|
1RP9
 
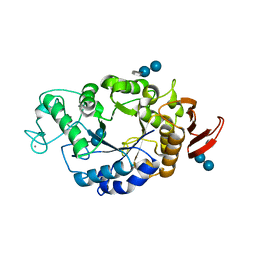 | | Crystal structure of barley alpha-amylase isozyme 1 (amy1) inactive mutant d180a in complex with acarbose | | 分子名称: | 4,6-dideoxy-4-{[(1S,5R,6S)-3-formyl-5,6-dihydroxy-4-oxocyclohex-2-en-1-yl]amino}-alpha-D-xylo-hex-5-enopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, 4,6-dideoxy-4-{[(1S,5R,6S)-3-formyl-5,6-dihydroxy-4-oxocyclohex-2-en-1-yl]amino}-alpha-D-xylo-hex-5-enopyranose-(1-4)-beta-D-glucopyranose-(1-4)-alpha-D-glucopyranose, Alpha-amylase type 1 isozyme, ... | | 著者 | Robert, X, Haser, R, Aghajari, N. | | 登録日 | 2003-12-03 | | 公開日 | 2005-06-07 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Oligosaccharide Binding to Barley {alpha}-Amylase 1
J.Biol.Chem., 280, 2005
|
|
1PTA
 
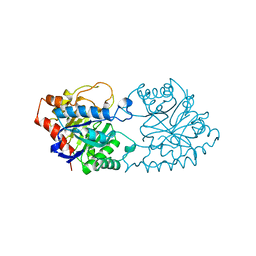 | |
1CET
 
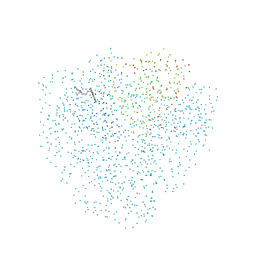 | | CHLOROQUINE BINDS IN THE COFACTOR BINDING SITE OF PLASMODIUM FALCIPARUM LACTATE DEHYDROGENASE. | | 分子名称: | N4-(7-CHLORO-QUINOLIN-4-YL)-N1,N1-DIETHYL-PENTANE-1,4-DIAMINE, PROTEIN (L-LACTATE DEHYDROGENASE) | | 著者 | Read, J.A, Wilkinson, K.W, Tranter, R, Sessions, R.B, Brady, R.L. | | 登録日 | 1999-03-10 | | 公開日 | 1999-03-19 | | 最終更新日 | 2023-08-09 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Chloroquine binds in the cofactor binding site of Plasmodium falciparum lactate dehydrogenase.
J.Biol.Chem., 274, 1999
|
|
1EQC
 
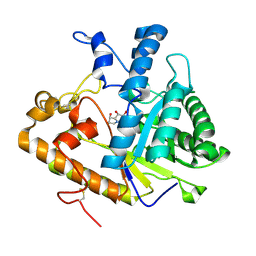 | | EXO-B-(1,3)-GLUCANASE FROM CANDIDA ALBICANS IN COMPLEX WITH CASTANOSPERMINE AT 1.85 A | | 分子名称: | CASTANOSPERMINE, EXO-(B)-(1,3)-GLUCANASE | | 著者 | Cutfield, S.M, Davies, G.J, Murshudov, G, Anderson, B.F, Moody, P.C.E, Sullivan, P.A, Cutfield, J.F. | | 登録日 | 2000-04-03 | | 公開日 | 2000-10-03 | | 最終更新日 | 2017-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | The structure of the exo-beta-(1,3)-glucanase from Candida albicans in native and bound forms: relationship between a pocket and groove in family 5 glycosyl hydrolases.
J.Mol.Biol., 294, 1999
|
|
2V6Y
 
 | | Structure of the MIT domain from a S. solfataricus Vps4-like ATPase | | 分子名称: | AAA FAMILY ATPASE, P60 KATANIN, S,R MESO-TARTARIC ACID | | 著者 | Obita, T, Saksena, S, Ghazi-Tabatabai, S, Gill, D.J, Perisic, O, Emr, S.D, Williams, R.L. | | 登録日 | 2007-07-24 | | 公開日 | 2007-10-16 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Structural Basis for Selective Recognition of Escrt-III by the Aaa ATPase Vps4
Nature, 449, 2007
|
|
2VGM
 
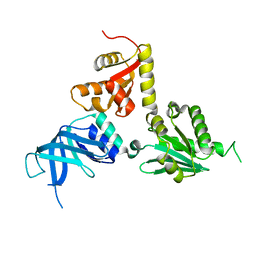 | |
2VGN
 
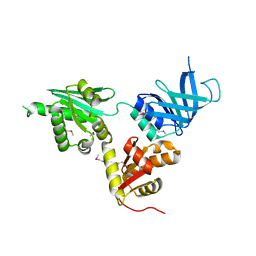 | | Structure of S. cerevisiae Dom34, a translation termination-like factor involved in RNA quality control pathways and interacting with Hbs1 (SelenoMet-labeled protein) | | 分子名称: | DOM34, GLYCEROL, PHOSPHATE ION | | 著者 | Graille, M, Chaillet, M, Van Tilbeurgh, H. | | 登録日 | 2007-11-14 | | 公開日 | 2008-01-22 | | 最終更新日 | 2021-03-17 | | 実験手法 | X-RAY DIFFRACTION (2.505 Å) | | 主引用文献 | Structure of Yeast Dom34: A Protein Related to Translation Termination Factor Erf1 and Involved in No-Go Decay.
J.Biol.Chem., 283, 2008
|
|
1WBK
 
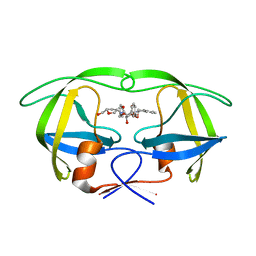 | |
1I6D
 
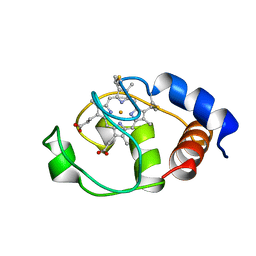 | | SOLUTION STRUCTURE OF THE FUNCTIONAL DOMAIN OF PARACOCCUS DENITRIFICANS CYTOCHROME C552 IN THE REDUCED STATE | | 分子名称: | CYTOCHROME C552, HEME C | | 著者 | Reincke, B, Perez, C, Pristovsek, P, Luecke, C, Ludwig, C, Loehr, F, Rogov, V.V, Ludwig, B, Rueterjans, H. | | 登録日 | 2001-03-02 | | 公開日 | 2001-10-17 | | 最終更新日 | 2022-02-23 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure and dynamics of the functional domain of Paracoccus denitrificans cytochrome c(552) in both redox states.
Biochemistry, 40, 2001
|
|
1CEQ
 
 | | CHLOROQUINE BINDS IN THE COFACTOR BINDING SITE OF PLASMODIUM FALCIPARUM LACTATE DEHYDROGENASE. | | 分子名称: | PROTEIN (L-LACTATE DEHYDROGENASE) | | 著者 | Read, J.A, Wilkinson, K.W, Tranter, R, Sessions, R.B, Brady, R.L. | | 登録日 | 1999-03-10 | | 公開日 | 1999-03-19 | | 最終更新日 | 2023-08-09 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Chloroquine binds in the cofactor binding site of Plasmodium falciparum lactate dehydrogenase.
J.Biol.Chem., 274, 1999
|
|
1GM9
 
 | | Crystal structures of penicillin acylase enzyme-substrate complexes: Structural insights into the catalytic mechanism | | 分子名称: | 1,2-ETHANEDIOL, CALCIUM ION, N-[(2S,4S,6R)-2-(DIHYDROXYMETHYL)-4-HYDROXY-3,3-DIMETHYL-7-OXO-4LAMBDA~4~-THIA-1-AZABICYCLO[3.2.0]HEPT-6-YL]-2-PHENYLAC ETAMIDE, ... | | 著者 | McVey, C.E, Walsh, M.A, Dodson, G.G, Wilson, K.S, Brannigan, J.A. | | 登録日 | 2001-09-12 | | 公開日 | 2001-11-28 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal Structures of Penicillin Acylase Enzyme- Substrate Complexes: Structural Insights Into the Catalytic Mechanism
J.Mol.Biol., 313, 2001
|
|
1GM8
 
 | | Crystal structures of penicillin acylase enzyme-substrate complexes: Structural insights into the catalytic mechanism | | 分子名称: | CALCIUM ION, N-[(2S,4S,6R)-2-(DIHYDROXYMETHYL)-4-HYDROXY-3,3-DIMETHYL-7-OXO-4LAMBDA~4~-THIA-1-AZABICYCLO[3.2.0]HEPT-6-YL]-2-PHENYLAC ETAMIDE, PENICILLIN G ACYLASE ALPHA SUBUNIT, ... | | 著者 | McVey, C.E, Walsh, M.A, Dodson, G.G, Wilson, K.S, Brannigan, J.A. | | 登録日 | 2001-09-11 | | 公開日 | 2001-11-28 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal Structures of Penicillin Acylase Enzyme- Substrate Complexes: Structural Insights Into the Catalytic Mechanism
J.Mol.Biol., 313, 2001
|
|
4HP0
 
 | | Crystal Structure of Hen Egg White Lysozyme in complex with GN3-M | | 分子名称: | 1-DEOXYNOJIRIMYCIN, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Lysozyme C | | 著者 | Umemoto, N, Numata, T, Ohnuma, T, Fukamizo, T. | | 登録日 | 2012-10-23 | | 公開日 | 2013-01-16 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.19 Å) | | 主引用文献 | A novel transition-state analogue for lysozyme, 4-O-beta-tri-N-acetylchitotriosyl moranoline, provided evidence supporting the covalent glycosyl-enzyme intermediate.
J.Biol.Chem., 288, 2013
|
|
1RP8
 
 | | Crystal structure of barley alpha-amylase isozyme 1 (amy1) inactive mutant d180a in complex with maltoheptaose | | 分子名称: | Alpha-amylase type 1 isozyme, CALCIUM ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, ... | | 著者 | Robert, X, Haser, R, Aghajari, N. | | 登録日 | 2003-12-03 | | 公開日 | 2005-06-07 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Oligosaccharide Binding to Barley {alpha}-Amylase 1
J.Biol.Chem., 280, 2005
|
|
