5B81
 
 | |
7B71
 
 | | Single modified phosphoryl guanidine DNA duplex, Sp diastereomer | | 分子名称: | DNA (5'-D(*CP*AP*CP*GP*CP*CP*GP*CP*TP*G)-3'), DNA (5'-D(*CP*AP*GP*CP*GP*GP*CP*GP*(SGT)P*G)-3') | | 著者 | Lomzov, A.A, Shernuykov, A.V, Apukhtina, V.S, Pyshnyi, D.V. | | 登録日 | 2020-12-09 | | 公開日 | 2021-01-13 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Single modified phosphoryl guanidine DNA duplex, Sp diastereomer
To Be Published
|
|
7VP1
 
 | |
7VP3
 
 | |
3ULD
 
 | |
3UKB
 
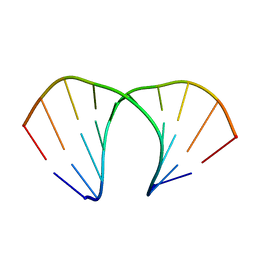 | | (R)-cEt-BNA decamer structure | | 分子名称: | DNA (5'-D(*GP*CP*GP*TP*AP*(RCE)P*AP*CP*GP*C)-3') | | 著者 | Pallan, P.S, Egli, M. | | 登録日 | 2011-11-09 | | 公開日 | 2012-06-20 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.42 Å) | | 主引用文献 | Structure and nuclease resistance of 2',4'-constrained 2'-O-methoxyethyl (cMOE) and 2'-O-ethyl (cEt) modified DNAs.
Chem.Commun.(Camb.), 48, 2012
|
|
1VPK
 
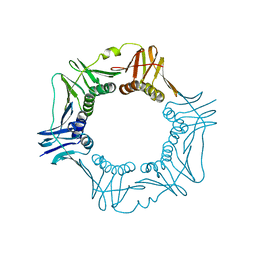 | |
2M2E
 
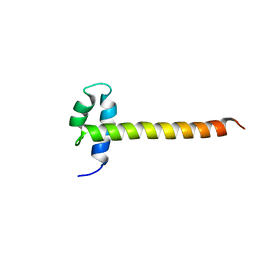 | | Solution NMR structure of the SANT domain of human DNAJC2, Northeast structural genomics consortium target HR8254a | | 分子名称: | DnaJ homolog subfamily C member 2 | | 著者 | Lemak, A, Yee, A, Houliston, S, Garcia, M, Ong, M, Montelione, G.T, Arrowsmith, C, Northeast Structural Genomics Consortium (NESG), Structural Genomics Consortium (SGC) | | 登録日 | 2012-12-18 | | 公開日 | 2013-02-20 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | NMR solution structure of the SANT domain of human DnaJC2.
To be Published
|
|
5CKK
 
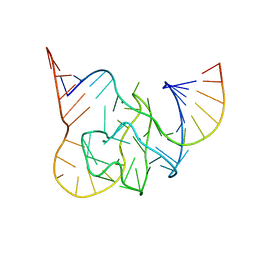 | |
8CS0
 
 | |
2VA8
 
 | | DNA Repair Helicase Hel308 | | 分子名称: | SKI2-TYPE HELICASE, SULFATE ION | | 著者 | Johnson, K.A, Richards, J, Liu, H, McMahon, S, Oke, M, Carter, L, Naismith, J.H, White, M.F. | | 登録日 | 2007-08-30 | | 公開日 | 2008-01-15 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structure of the DNA Repair Helicase Hel308 Reveals DNA Binding and Autoinhibitory Domains.
J.Biol.Chem., 283, 2008
|
|
5ZTJ
 
 | | Crystal Structure of GyraseA C-Terminal Domain from Salmonella typhi at 2.4A Resolution | | 分子名称: | DNA gyrase subunit A | | 著者 | Sachdeva, E, Gupta, D, Tiwari, P, Kaur, G, Sharma, S, Singh, T.P, Ethayathulla, A.S, Kaur, P. | | 登録日 | 2018-05-03 | | 公開日 | 2019-05-15 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | The pivot point arginines identified in the beta-pinwheel structure of C-terminal domain from Salmonella Typhi DNA Gyrase A subunit.
Sci Rep, 10, 2020
|
|
6A28
 
 | | Crystal structure of PprA W183R mutant form 2 | | 分子名称: | DNA repair protein PprA, SULFATE ION | | 著者 | Adachi, M, Shibazaki, C, Shimizu, R, Arai, S, Satoh, K, Narumi, I, Kuroki, R. | | 登録日 | 2018-06-09 | | 公開日 | 2018-12-26 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.193 Å) | | 主引用文献 | Extended structure of pleiotropic DNA repair-promoting protein PprA from Deinococcus radiodurans.
FASEB J., 33, 2019
|
|
1VT8
 
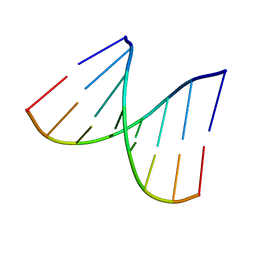 | | Crystal structure of D(GGGCGCCC)-hexagonal form | | 分子名称: | DNA (5'-D(*GP*GP*GP*CP*GP*CP*CP*C)-3') | | 著者 | Shakked, Z, Guerstein-Guzikevich, G, Eisenstein, M, Frolow, F, Rabinovich, D. | | 登録日 | 1996-12-02 | | 公開日 | 2011-07-13 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | The conformation of the DNA double helix in the crystal is dependent on its environment.
Nature, 342, 1989
|
|
1NG9
 
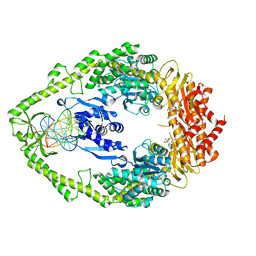 | | E.coli MutS R697A: an ATPase-asymmetry mutant | | 分子名称: | 5'-D(*AP*GP*CP*TP*GP*CP*CP*AP*GP*GP*CP*AP*CP*CP*AP*GP*TP*GP*TP*CP*AP*GP*CP*GP*TP*CP*CP*TP*AP*T)-3', 5'-D(*AP*TP*AP*GP*GP*AP*CP*GP*CP*TP*GP*AP*CP*AP*CP*TP*GP*GP*TP*GP*CP*TP*TP*GP*GP*CP*AP*GP*CP*T)-3', ADENOSINE-5'-DIPHOSPHATE, ... | | 著者 | Lamers, M.H, Winterwerp, H.H.K, Sixma, T.K. | | 登録日 | 2002-12-17 | | 公開日 | 2003-02-11 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | The alternating ATPase domains of MutS control DNA mismatch repair
Embo J., 22, 2003
|
|
4QFQ
 
 | |
2V1U
 
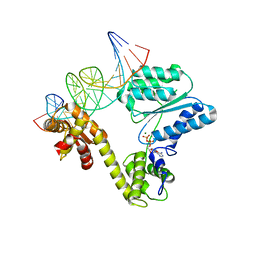 | | STRUCTURE OF THE AEROPYRUM PERNIX ORC1 PROTEIN IN COMPLEX WITH DNA | | 分子名称: | 5'-D(*AP*CP*CP*CP*CP*TP*CP*CP*GP*TP *TP*TP*CP*CP*TP*GP*TP*GP*GP*AP*GP*A)-3', 5'-D(*TP*CP*TP*CP*CP*AP*CP*AP*GP*GP *AP*AP*AP*CP*GP*GP*AP*GP*GP*GP*GP*T)-3', ADENOSINE-5'-DIPHOSPHATE, ... | | 著者 | Gaudier, M, Schuwirth, B.S, Westcott, S.L, Wigley, D.B. | | 登録日 | 2007-05-30 | | 公開日 | 2007-09-18 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Structural Basis of DNA Replication Origin Recognition by an Orc Protein.
Science, 317, 2007
|
|
1XF2
 
 | | Structure of Fab DNA-1 complexed with dT3 | | 分子名称: | 5'-D(*TP*TP*T)-3', SULFATE ION, antibody heavy chain Fab, ... | | 著者 | Schuermann, J.P, Prewitt, S.P, Deutscher, S.L, Tanner, J.J. | | 登録日 | 2004-09-13 | | 公開日 | 2005-04-12 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Evidence for Structural Plasticity of Heavy Chain Complementarity-determining Region 3 in Antibody-ssDNA Recognition
J.Mol.Biol., 347, 2005
|
|
1B22
 
 | | RAD51 (N-TERMINAL DOMAIN) | | 分子名称: | DNA REPAIR PROTEIN RAD51 | | 著者 | Aihara, H, Ito, Y, Kurumizaka, H, Yokoyama, S, Shibata, T, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 1998-12-04 | | 公開日 | 1999-12-03 | | 最終更新日 | 2023-12-27 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The N-terminal domain of the human Rad51 protein binds DNA: structure and a DNA binding surface as revealed by NMR.
J.Mol.Biol., 290, 1999
|
|
251D
 
 | |
6ABO
 
 | | human XRCC4 and IFFO1 complex | | 分子名称: | DNA repair protein XRCC4, GLYCEROL, Intermediate filament family orphan 1, ... | | 著者 | Li, J, Liu, L, Liang, H, Liu, Y, Xu, D. | | 登録日 | 2018-07-23 | | 公開日 | 2019-07-31 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.65 Å) | | 主引用文献 | The nucleoskeleton protein IFFO1 immobilizes broken DNA and suppresses chromosome translocation during tumorigenesis.
Nat.Cell Biol., 21, 2019
|
|
1XF3
 
 | | Structure of ligand-free Fab DNA-1 in space group P65 | | 分子名称: | Fab Light chain, Fab heavy chain | | 著者 | Schuermann, J.P, Prewitt, S.P, Deutscher, S.L, Tanner, J.J. | | 登録日 | 2004-09-13 | | 公開日 | 2005-04-12 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Evidence for Structural Plasticity of Heavy Chain Complementarity-determining Region 3 in Antibody-ssDNA Recognition
J.Mol.Biol., 347, 2005
|
|
7KWL
 
 | |
5J05
 
 | |
1XF4
 
 | | Structure of ligand-free Fab DNA-1 in space group P321 solved from crystals with perfect hemihedral twinning | | 分子名称: | Fab heavy chain, Fab light chain, SULFATE ION | | 著者 | Schuermann, J.P, Prewitt, S.P, Deutscher, S.L, Tanner, J.J. | | 登録日 | 2004-09-13 | | 公開日 | 2005-04-12 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Evidence for Structural Plasticity of Heavy Chain Complementarity-determining Region 3 in Antibody-ssDNA Recognition
J.Mol.Biol., 347, 2005
|
|
