5O8W
 
 | | CRYSTAL STRUCTURE ANALYSIS OF THE YEAST ELONGATION FACTOR COMPLEX EEF1A:EEF1BA | | 分子名称: | Elongation factor 1-alpha, Elongation factor 1-beta, GLUTAMINE, ... | | 著者 | Wirth, C, Andersen, G.R, Hunte, C. | | 登録日 | 2017-06-14 | | 公開日 | 2017-08-23 | | 最終更新日 | 2019-07-17 | | 実験手法 | X-RAY DIFFRACTION (1.67 Å) | | 主引用文献 | Protein glutaminylation is a yeast-specific posttranslational modification of elongation factor 1A.
J. Biol. Chem., 292, 2017
|
|
7O0G
 
 | |
6WQH
 
 | | Molecular basis for the ATPase-powered substrate translocation by the Lon AAA+ protease | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Ig2 substrate, Lon protease, ... | | 著者 | Zhang, K, Li, S, Hsiehb, K, Sub, S, Pintilie, G, Chiu, W, Chang, C. | | 登録日 | 2020-04-28 | | 公開日 | 2021-06-09 | | 最終更新日 | 2021-11-17 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Molecular basis for ATPase-powered substrate translocation by the Lon AAA+ protease.
J.Biol.Chem., 297, 2021
|
|
4X9E
 
 | | DEOXYGUANOSINETRIPHOSPHATE TRIPHOSPHOHYDROLASE from Escherichia coli with two DNA effector molecules | | 分子名称: | Deoxyguanosinetriphosphate triphosphohydrolase, MAGNESIUM ION, RNA (5'-R(P*CP*CP*C)-3') | | 著者 | Singh, D, Gawel, D, Itsko, M, Krahn, J.M, London, R.E, Schaaper, R.M. | | 登録日 | 2014-12-11 | | 公開日 | 2015-02-25 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Structure of Escherichia coli dGTP Triphosphohydrolase: A HEXAMERIC ENZYME WITH DNA EFFECTOR MOLECULES.
J.Biol.Chem., 290, 2015
|
|
7ZM7
 
 | | CryoEM structure of mitochondrial complex I from Chaetomium thermophilum (inhibited by DDM) | | 分子名称: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, Acyl carrier protein, ... | | 著者 | Laube, E, Kuehlbrandt, W. | | 登録日 | 2022-04-19 | | 公開日 | 2022-11-30 | | 最終更新日 | 2022-12-07 | | 実験手法 | ELECTRON MICROSCOPY (2.77 Å) | | 主引用文献 | Conformational changes in mitochondrial complex I of the thermophilic eukaryote Chaetomium thermophilum.
Sci Adv, 8, 2022
|
|
7ZMG
 
 | | CryoEM structure of mitochondrial complex I from Chaetomium thermophilum (state 1) | | 分子名称: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, Acyl carrier protein, ... | | 著者 | Laube, E, Kuehlbrandt, W. | | 登録日 | 2022-04-19 | | 公開日 | 2022-11-30 | | 最終更新日 | 2022-12-07 | | 実験手法 | ELECTRON MICROSCOPY (2.44 Å) | | 主引用文献 | Conformational changes in mitochondrial complex I of the thermophilic eukaryote Chaetomium thermophilum.
Sci Adv, 8, 2022
|
|
7ZMB
 
 | | CryoEM structure of mitochondrial complex I from Chaetomium thermophilum (state 2) | | 分子名称: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, Acyl carrier protein, ... | | 著者 | Laube, E, Kuehlbrandt, W. | | 登録日 | 2022-04-19 | | 公開日 | 2022-11-30 | | 最終更新日 | 2022-12-07 | | 実験手法 | ELECTRON MICROSCOPY (2.75 Å) | | 主引用文献 | Conformational changes in mitochondrial complex I of the thermophilic eukaryote Chaetomium thermophilum.
Sci Adv, 8, 2022
|
|
3CNJ
 
 | |
4P1W
 
 | |
4WHB
 
 | | Crystal structure of phenylurea hydrolase B | | 分子名称: | Phenylurea hydrolase B, ZINC ION | | 著者 | Sugrue, E, Carr, P.D, Khurana, J.L, Jackson, C.J. | | 登録日 | 2014-09-21 | | 公開日 | 2015-02-11 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.958 Å) | | 主引用文献 | Evolutionary Expansion of the Amidohydrolase Superfamily in Bacteria in Response to the Synthetic Compounds Molinate and Diuron.
Appl.Environ.Microbiol., 81, 2015
|
|
4WGX
 
 | | Crystal Structure of Molinate Hydrolase | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, COBALT (II) ION, Molinate hydrolase | | 著者 | Sugrue, E, Carr, P.D, Fraser, N.J, Hopkins, D.H, Jackson, C.J. | | 登録日 | 2014-09-19 | | 公開日 | 2015-02-11 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.29 Å) | | 主引用文献 | Evolutionary Expansion of the Amidohydrolase Superfamily in Bacteria in Response to the Synthetic Compounds Molinate and Diuron.
Appl.Environ.Microbiol., 81, 2015
|
|
6WAY
 
 | | C-terminal SH2 domain of p120RasGAP in complex with p190RhoGAP phosphotyrosine peptide | | 分子名称: | Ras GTPase-activating protein 1, Rho GTPase-activating protein 35 | | 著者 | Jaber Chehayeb, R, Wang, J, Stiegler, A.L, Boggon, T.J. | | 登録日 | 2020-03-26 | | 公開日 | 2020-06-17 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | The GTPase-activating protein p120RasGAP has an evolutionarily conserved "FLVR-unique" SH2 domain.
J.Biol.Chem., 295, 2020
|
|
3CNR
 
 | |
2RMN
 
 | |
3CU7
 
 | | Human Complement Component 5 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CADMIUM ION, ... | | 著者 | Fredslund, F, Andersen, G.R. | | 登録日 | 2008-04-16 | | 公開日 | 2008-06-10 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (3.105 Å) | | 主引用文献 | Structure of and influence of a tick complement inhibitor on human complement component 5
Nat.Immunol., 9, 2008
|
|
3DDL
 
 | | Crystallographic Structure of Xanthorhodopsin, a Light-Driven Ion Pump with Dual Chromophore | | 分子名称: | 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, RETINAL, ... | | 著者 | Stagno, J, Luecke, H, Schobert, B, Lanyi, J.K, Imasheva, E.S, Wang, J.M, Balashov, S.P. | | 登録日 | 2008-06-05 | | 公開日 | 2008-10-14 | | 最終更新日 | 2016-06-08 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystallographic structure of xanthorhodopsin, the light-driven proton pump with a dual chromophore.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
7ZMH
 
 | | CryoEM structure of mitochondrial complex I from Chaetomium thermophilum (state 1) - membrane arm | | 分子名称: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, Acyl carrier protein, ... | | 著者 | Laube, E, Kuehlbrandt, W. | | 登録日 | 2022-04-19 | | 公開日 | 2022-11-30 | | 最終更新日 | 2022-12-07 | | 実験手法 | ELECTRON MICROSCOPY (2.47 Å) | | 主引用文献 | Conformational changes in mitochondrial complex I of the thermophilic eukaryote Chaetomium thermophilum.
Sci Adv, 8, 2022
|
|
7ZM8
 
 | | CryoEM structure of mitochondrial complex I from Chaetomium thermophilum (inhibited by DDM) - membrane arm | | 分子名称: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, Acyl carrier protein, ... | | 著者 | Laube, E, Kuehlbrandt, W. | | 登録日 | 2022-04-19 | | 公開日 | 2022-11-30 | | 最終更新日 | 2022-12-07 | | 実験手法 | ELECTRON MICROSCOPY (2.76 Å) | | 主引用文献 | Conformational changes in mitochondrial complex I of the thermophilic eukaryote Chaetomium thermophilum.
Sci Adv, 8, 2022
|
|
7ZME
 
 | | CryoEM structure of mitochondrial complex I from Chaetomium thermophilum (state 2) - membrane arm | | 分子名称: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, Acyl carrier protein, ... | | 著者 | Laube, E, Kuehlbrandt, W. | | 登録日 | 2022-04-19 | | 公開日 | 2022-11-30 | | 最終更新日 | 2022-12-07 | | 実験手法 | ELECTRON MICROSCOPY (2.83 Å) | | 主引用文献 | Conformational changes in mitochondrial complex I of the thermophilic eukaryote Chaetomium thermophilum.
Sci Adv, 8, 2022
|
|
1E9L
 
 | |
3UR3
 
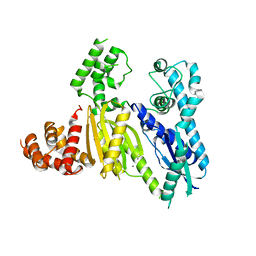 | | Structure of the Cmr2 subunit of the CRISPR RNA silencing complex | | 分子名称: | CALCIUM ION, Cmr2dHD, ZINC ION | | 著者 | Cocozaki, A.I, Ramia, N.F, Shao, Y, Hale, C.R, Terns, R.M, Terns, M.P, Li, H. | | 登録日 | 2011-11-21 | | 公開日 | 2012-03-21 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.405 Å) | | 主引用文献 | Structure of the Cmr2 Subunit of the CRISPR-Cas RNA Silencing Complex.
Structure, 20, 2012
|
|
5WAN
 
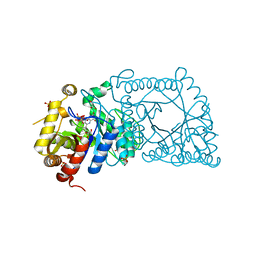 | | Crystal Structure of a flavoenzyme RutA in the pyrimidine catabolic pathway | | 分子名称: | FLAVIN MONONUCLEOTIDE, GLYCEROL, Pyrimidine monooxygenase RutA, ... | | 著者 | Zhang, Y, Mukherjee, T, Abdelwahed, S, Begley, T.P, Ealick, S.E. | | 登録日 | 2017-06-26 | | 公開日 | 2017-07-19 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.798 Å) | | 主引用文献 | Catalysis of a flavoenzyme-mediated amide hydrolysis.
J. Am. Chem. Soc., 132, 2010
|
|
3LTN
 
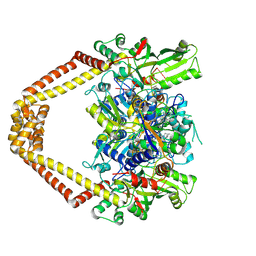 | | Inhibitor-stabilized topoisomerase IV-DNA cleavage complex (S. pneumoniae) | | 分子名称: | 3-amino-7-{(3R)-3-[(1S)-1-aminoethyl]pyrrolidin-1-yl}-1-cyclopropyl-6-fluoro-8-methylquinazoline-2,4(1H,3H)-dione, 5'-D(*AP*CP*CP*AP*AP*GP*GP*T*CP*AP*TP*GP*AP*AP*T)-3', 5'-D(*CP*TP*GP*TP*TP*TP*TP*A*CP*GP*TP*GP*CP*AP*T)-3', ... | | 著者 | Laponogov, I, Pan, X.-S, Veselkov, D.A, McAuley, K.E, Fisher, L.M, Sanderson, M.R. | | 登録日 | 2010-02-16 | | 公開日 | 2010-05-05 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Structural Basis of Gate-DNA Breakage and Resealing by Type II Topoisomerases
Plos One, 5, 2010
|
|
521P
 
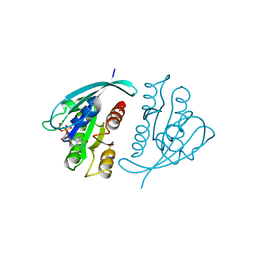 | | THREE-DIMENSIONAL STRUCTURES OF H-RAS P21 MUTANTS: MOLECULAR BASIS FOR THEIR INABILITY TO FUNCTION AS SIGNAL SWITCH MOLECULES | | 分子名称: | GUANOSINE-5'-TRIPHOSPHATE, H-RAS P21 PROTEIN, MAGNESIUM ION | | 著者 | Schlichting, I, Krengel, U, Kabsch, W, Wittinghofer, A, Pai, E.F. | | 登録日 | 1991-06-06 | | 公開日 | 1994-01-31 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Three-dimensional structures of H-ras p21 mutants: molecular basis for their inability to function as signal switch molecules.
Cell(Cambridge,Mass.), 62, 1990
|
|
5NTT
 
 | | Crystal structure of human Mps1 (TTK) C604Y mutant in complex with NMS-P715 | | 分子名称: | 1,2-ETHANEDIOL, Dual specificity protein kinase TTK, N-(2,6-DIETHYLPHENYL)-1-METHYL-8-({4-[(1-METHYLPIPERIDIN-4-YL)CARBAMOYL]-2-(TRIFLUOROMETHOXY)PHENYL}AMINO)-4,5-DIHYDRO-1H-PYRAZOLO[4,3-H]QUINAZOLINE-3-CARBOXAMIDE | | 著者 | Hiruma, Y, Joosten, R.P, Perrakis, A. | | 登録日 | 2017-04-28 | | 公開日 | 2017-07-26 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.75 Å) | | 主引用文献 | Understanding inhibitor resistance in Mps1 kinase through novel biophysical assays and structures.
J. Biol. Chem., 292, 2017
|
|
