8AIH
 
 | | Crystal Structure of Enterococcus faecium Nicotinate Nucleotide Adenylyltransferase at 1.9 Angstroms Resolution | | 分子名称: | DIHYDROGENPHOSPHATE ION, Probable nicotinate-nucleotide adenylyltransferase, SULFATE ION | | 著者 | Pandian, R, Jeje, O.A, Sayed, Y, Achilonu, I.A. | | 登録日 | 2022-07-26 | | 公開日 | 2023-08-16 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Obtaining high yield recombinant Enterococcus faecium nicotinate nucleotide adenylyltransferase for X-ray crystallography and biophysical studies.
Int.J.Biol.Macromol., 250, 2023
|
|
8AII
 
 | | High Resolution Crystal Structure of Enterococcus faecium Nicotinate Nucleotide Adenylyltransferase Complexed with Adenine | | 分子名称: | ADENINE, MAGNESIUM ION, Probable nicotinate-nucleotide adenylyltransferase, ... | | 著者 | Pandian, R, Jeje, O.A, Sayed, Y, Achilonu, I.A. | | 登録日 | 2022-07-26 | | 公開日 | 2023-08-16 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (1.82 Å) | | 主引用文献 | Obtaining high yield recombinant Enterococcus faecium nicotinate nucleotide adenylyltransferase for X-ray crystallography and biophysical studies.
Int.J.Biol.Macromol., 250, 2023
|
|
8ALS
 
 | |
8BHZ
 
 | |
6K4J
 
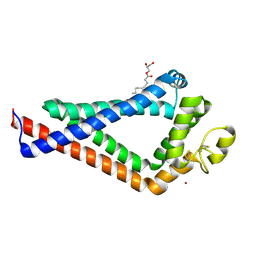 | | Crystal Structure of the the CD9 | | 分子名称: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CD9 antigen, NICKEL (II) ION, ... | | 著者 | Umeda, R, Nishizawa, T, Sato, K, Nureki, O. | | 登録日 | 2019-05-24 | | 公開日 | 2020-05-13 | | 実験手法 | X-RAY DIFFRACTION (2.701 Å) | | 主引用文献 | Structural insights into tetraspanin CD9 function.
Nat Commun, 11, 2020
|
|
1MI7
 
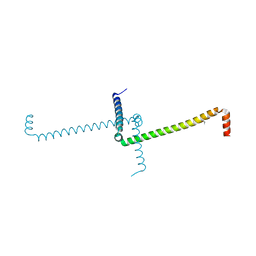 | | Crystal Structure of Domain Swapped trp Aporepressor in 30%(v/v) Isopropanol | | 分子名称: | ISOPROPYL ALCOHOL, Trp operon repressor | | 著者 | Lawson, C.L, Benoff, B, Berger, T, Berman, H.M, Carey, J. | | 登録日 | 2002-08-22 | | 公開日 | 2003-09-02 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | E. coli trp repressor forms a domain-swapped array in aqueous alcohol.
Structure, 12, 2004
|
|
5QYI
 
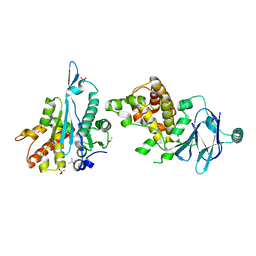 | | PanDDA analysis group deposition -- Aar2/RNaseH in complex with fragment F2X-Entry G12a | | 分子名称: | A1 cistron-splicing factor AAR2, Pre-mRNA-splicing factor 8, R-1,2-PROPANEDIOL, ... | | 著者 | Weiss, M.S, Wollenhaupt, J, Metz, A, Barthel, T, Lima, G.M.A, Heine, A, Mueller, U, Klebe, G. | | 登録日 | 2020-02-12 | | 公開日 | 2020-06-10 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.58 Å) | | 主引用文献 | F2X-Universal and F2X-Entry: Structurally Diverse Compound Libraries for Crystallographic Fragment Screening.
Structure, 28, 2020
|
|
3Q3B
 
 | |
7FR2
 
 | |
7FR3
 
 | |
7FRC
 
 | |
7FRD
 
 | |
6JL2
 
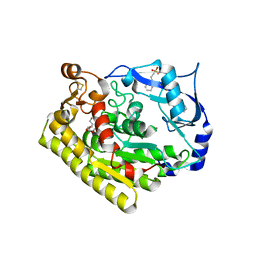 | |
2GYW
 
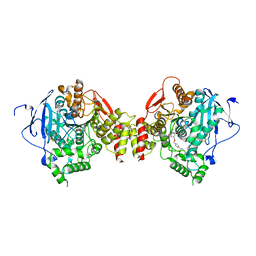 | | Crystal Structure of Mus musculus Acetylcholinesterase in Complex with Obidoxime | | 分子名称: | 1,1'-(OXYDIMETHYLENE)BIS(4-FORMYLPYRIDINIUM)DIOXIME, 2-acetamido-2-deoxy-beta-D-glucopyranose, Acetylcholinesterase, ... | | 著者 | Pang, Y.P, Boman, M, Artursson, E, Akfur, C, Lundberg, S. | | 登録日 | 2006-05-10 | | 公開日 | 2006-08-15 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Crystal structures of acetylcholinesterase in complex with HI-6, Ortho-7 and obidoxime: Structural basis for differences in the ability to reactivate tabun conjugates.
Biochem.Pharm., 72, 2006
|
|
5J8T
 
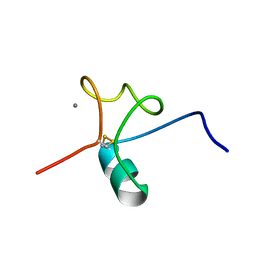 | |
2GYV
 
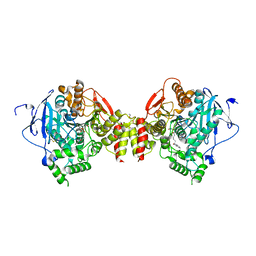 | | Crystal structure of Mus musculus Acetylcholinesterase in complex with Ortho-7 | | 分子名称: | 1,7-HEPTYLENE-BIS-N,N'-SYN-2-PYRIDINIUMALDOXIME, 2-acetamido-2-deoxy-beta-D-glucopyranose, 3,6,9,12,15-PENTAOXAHEPTADECANE, ... | | 著者 | Pang, Y.P, Boman, M, Artursson, E, Akfur, C, Lundberg, S. | | 登録日 | 2006-05-10 | | 公開日 | 2006-08-15 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Crystal structures of acetylcholinesterase in complex with HI-6, Ortho-7 and obidoxime: Structural basis for differences in the ability to reactivate tabun conjugates.
Biochem.Pharm., 72, 2006
|
|
2HPW
 
 | | Green fluorescent protein from Clytia gregaria | | 分子名称: | Green fluorescent protein | | 著者 | Stepanyuk, G, Liu, Z.J, Vysotski, S.E, Lee, J, Rose, J.P, Wang, B.C, Southeast Collaboratory for Structural Genomics (SECSG) | | 登録日 | 2006-07-17 | | 公開日 | 2006-09-12 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Crystal Structure of Green Fluorescent Protein from Clytia Gregaria at 1.55 A resolution
To be Published
|
|
2GYU
 
 | | Crystal structure of Mus musculus Acetylcholinesterase in complex with HI-6 | | 分子名称: | 1-ETHOXY-2-(2-ETHOXYETHOXY)ETHANE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(AMINOCARBONYL)-1-[({2-[(E)-(HYDROXYIMINO)METHYL]PYRIDINIUM-1-YL}METHOXY)METHYL]PYRIDINIUM, ... | | 著者 | Pang, Y.P, Boman, M, Artursson, E, Akfur, C, Lundberg, S. | | 登録日 | 2006-05-10 | | 公開日 | 2006-08-15 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Crystal structures of acetylcholinesterase in complex with HI-6, Ortho-7 and obidoxime: Structural basis for differences in the ability to reactivate tabun conjugates.
Biochem.Pharm., 72, 2006
|
|
2FF1
 
 | | Crystal structure of Trypanosoma vivax nucleoside hydrolase soaked with ImmucillinH | | 分子名称: | 1,4-DIDEOXY-4-AZA-1-(S)-(9-DEAZAHYPOXANTHIN-9-YL)-D-RIBITOL, CALCIUM ION, IAG-nucleoside hydrolase | | 著者 | Versees, W, Barlow, J, Steyaert, J. | | 登録日 | 2005-12-18 | | 公開日 | 2006-05-23 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (2.07 Å) | | 主引用文献 | Transition-state Complex of the Purine-specific Nucleoside Hydrolase of T.vivax: Enzyme Conformational Changes and Implications for Catalysis.
J.Mol.Biol., 359, 2006
|
|
2FF2
 
 | | Crystal structure of Trypanosoma vivax nucleoside hydrolase co-crystallized with ImmucillinH | | 分子名称: | 1,4-DIDEOXY-4-AZA-1-(S)-(9-DEAZAHYPOXANTHIN-9-YL)-D-RIBITOL, CALCIUM ION, IAG-nucleoside hydrolase, ... | | 著者 | Versees, W, Barlow, J, Steyaert, J. | | 登録日 | 2005-12-18 | | 公開日 | 2006-05-23 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Transition-state Complex of the Purine-specific Nucleoside Hydrolase of T.vivax: Enzyme Conformational Changes and Implications for Catalysis.
J.Mol.Biol., 359, 2006
|
|
3RED
 
 | | 3.0 A structure of the Prunus mume hydroxynitrile lyase isozyme-1 | | 分子名称: | FLAVIN-ADENINE DINUCLEOTIDE, Hydroxynitrile lyase | | 著者 | Cielo, C.B.C, Yamane, T, Asano, Y, Watanabe, N, Suzuki, A, Fukuta, Y. | | 登録日 | 2011-04-04 | | 公開日 | 2012-06-20 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (3.03 Å) | | 主引用文献 | Crystal Structure of a native FAD-dependent Hydroxynitrile Lyase derived from the Japanese apricot, Prunus mume
To be Published
|
|
3TWZ
 
 | | Phosphorylated Bacillus cereus phosphopentomutase in space group P212121 | | 分子名称: | MANGANESE (II) ION, Phosphopentomutase | | 著者 | Panosian, T.P, Nanneman, D.P, Bachmann, B.O, Iverson, T.M. | | 登録日 | 2011-09-22 | | 公開日 | 2012-02-29 | | 最終更新日 | 2012-03-21 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Molecular Differences between a Mutase and a Phosphatase: Investigations of the Activation Step in Bacillus cereus Phosphopentomutase.
Biochemistry, 51, 2012
|
|
3TX0
 
 | | Unphosphorylated Bacillus cereus phosphopentomutase in a P212121 crystal form | | 分子名称: | MANGANESE (II) ION, Phosphopentomutase | | 著者 | Panosian, T.P, Nanneman, D.P, Bachmann, B.O, Iverson, T.M. | | 登録日 | 2011-09-22 | | 公開日 | 2012-02-29 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.26 Å) | | 主引用文献 | Molecular Differences between a Mutase and a Phosphatase: Investigations of the Activation Step in Bacillus cereus Phosphopentomutase.
Biochemistry, 51, 2012
|
|
2LMA
 
 | | Solution structure of CD4+ T cell derived peptide Thp5 | | 分子名称: | Thp5 peptide | | 著者 | Pandey, N.K, Khan, M.M, Chatterjee, S, Dwivedi, V.P, Tousif, S, Das, J, Van Kaer, L. | | 登録日 | 2011-11-29 | | 公開日 | 2011-12-28 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | CD4+ T cell derived novel peptide Thp5 induces IL-4 production in CD4+ T cells to direct T helper 2 cell differentiation
J.Biol.Chem., 2011
|
|
3QPN
 
 | | Structure of PDE10-inhibitor complex | | 分子名称: | 6-methoxy-7-[2-(quinolin-2-yl)ethoxy]quinazoline, MAGNESIUM ION, SULFATE ION, ... | | 著者 | Pandit, J, Marr, E.S. | | 登録日 | 2011-02-14 | | 公開日 | 2011-06-15 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Use of Structure-Based Design to Discover a Potent, Selective, In Vivo Active Phosphodiesterase 10A Inhibitor Lead Series for the Treatment of Schizophrenia.
J.Med.Chem., 54, 2011
|
|
