5QFZ
 
 | | PanDDA analysis group deposition -- Crystal structure of PTP1B in complex with compound_FMOPL000711a | | 分子名称: | (4-chloranyl-2-methyl-pyrazol-3-yl)-piperidin-1-yl-methanone, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Tyrosine-protein phosphatase non-receptor type 1 | | 著者 | Keedy, D.A, Hill, Z.B, Biel, J.T, Kang, E, Rettenmaier, T.J, Brandao-Neto, J, von Delft, F, Wells, J.A, Fraser, J.S. | | 登録日 | 2018-08-30 | | 公開日 | 2018-10-10 | | 最終更新日 | 2024-11-20 | | 実験手法 | X-RAY DIFFRACTION (1.723 Å) | | 主引用文献 | An expanded allosteric network in PTP1B by multitemperature crystallography, fragment screening, and covalent tethering.
Elife, 7, 2018
|
|
5YPP
 
 | | Crystal structure of IlvN.Val-1a | | 分子名称: | ACETATE ION, Acetolactate synthase isozyme 1 small subunit, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Sarma, S.P, Bansal, A, Schindelin, H, Demeler, B. | | 登録日 | 2017-11-02 | | 公開日 | 2018-09-19 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystallographic Structures of IlvN·Val/Ile Complexes: Conformational Selectivity for Feedback Inhibition of Aceto Hydroxy Acid Synthases.
Biochemistry, 58, 2019
|
|
5QFM
 
 | | PanDDA analysis group deposition -- Crystal structure of PTP1B in complex with compound_FMOMB000269a | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, N-[(1R,2S,4S)-bicyclo[2.2.1]heptan-2-yl]-N'-[(2R)-2-hydroxypropyl]thiourea, Tyrosine-protein phosphatase non-receptor type 1 | | 著者 | Keedy, D.A, Hill, Z.B, Biel, J.T, Kang, E, Rettenmaier, T.J, Brandao-Neto, J, von Delft, F, Wells, J.A, Fraser, J.S. | | 登録日 | 2018-08-30 | | 公開日 | 2018-10-10 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.83 Å) | | 主引用文献 | An expanded allosteric network in PTP1B by multitemperature crystallography, fragment screening, and covalent tethering.
Elife, 7, 2018
|
|
6LPF
 
 | | The crystal structure of human cytoplasmic LRS | | 分子名称: | 2'-(L-NORVALYL)AMINO-2'-DEOXYADENOSINE, 5'-O-(L-leucylsulfamoyl)adenosine, GLYCEROL, ... | | 著者 | Liu, R.J, Long, T, Li, H, Li, J, Zhao, J.H, Lin, J.Z, Palencia, A, Wang, M.Z, Cusack, S, Wang, E.D. | | 登録日 | 2020-01-10 | | 公開日 | 2020-03-25 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.49 Å) | | 主引用文献 | Molecular basis of the multifaceted functions of human leucyl-tRNA synthetase in protein synthesis and beyond.
Nucleic Acids Res., 48, 2020
|
|
3G7W
 
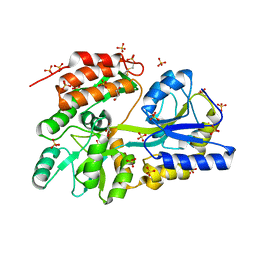 | | Islet Amyloid Polypeptide (IAPP or Amylin) Residues 1 to 22 fused to Maltose Binding Protein | | 分子名称: | GLYCEROL, Maltose-binding periplasmic protein, Islet amyloid polypeptide fusion protein, ... | | 著者 | Wiltzius, J.J.W, Sawaya, M.R, Eisenberg, D. | | 登録日 | 2009-02-11 | | 公開日 | 2009-06-23 | | 最終更新日 | 2024-11-27 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Atomic structures of IAPP (amylin) fusions suggest a mechanism for fibrillation and the role of insulin in the process
Protein Sci., 18, 2009
|
|
6BEI
 
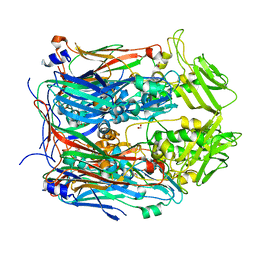 | | Crystal structure of VACV D13 in its apo (unbound) form | | 分子名称: | 1,2-ETHANEDIOL, FORMIC ACID, Scaffold protein D13 | | 著者 | Garriga, D, Accurso, C, Coulibaly, F. | | 登録日 | 2017-10-25 | | 公開日 | 2018-07-18 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.81 Å) | | 主引用文献 | Structural basis for the inhibition of poxvirus assembly by the antibiotic rifampicin.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5TM4
 
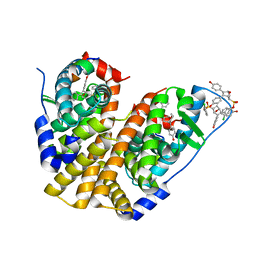 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in Complex with the OBHS-ASC Analog, 5-(4-((1R,4S,6R)-6-((3-chlorophenoxy)sulfonyl)-3-(4-hydroxyphenyl)-7-oxabicyclo[2.2.1]hept-2-en-2-yl)phenoxy)pentanoic acid | | 分子名称: | 5-{4-[(1S,4S,6R)-6-[(3-chlorophenoxy)sulfonyl]-3-(4-hydroxyphenyl)-7-oxabicyclo[2.2.1]hept-2-en-2-yl]phenoxy}pentanoic acid, Estrogen receptor, Nuclear receptor coactivator 2 | | 著者 | Nwachukwu, J.C, Srinivasan, S, Bruno, N.E, Nowak, J, Kojetin, D.J, Elemento, O, Katzenellenbogen, J.A, Nettles, K.W. | | 登録日 | 2016-10-12 | | 公開日 | 2017-01-18 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Systems Structural Biology Analysis of Ligand Effects on ER alpha Predicts Cellular Response to Environmental Estrogens and Anti-hormone Therapies.
Cell Chem Biol, 24, 2017
|
|
5TMM
 
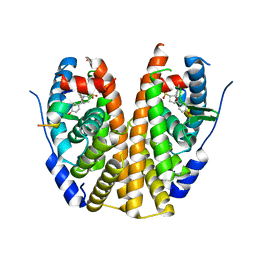 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in Complex with the OBHS-ASC analog, (E)-6-(4-((1R,4S,6R)-6-((4-bromophenoxy)sulfonyl)-3-(4-hydroxyphenyl)-7-oxabicyclo[2.2.1]hept-2-en-2-yl)phenyl)hex-5-enoic acid | | 分子名称: | 6-{4-[(1S,4S,6S)-6-[(4-bromophenoxy)sulfonyl]-3-(4-hydroxyphenyl)-7-oxabicyclo[2.2.1]hept-2-en-2-yl]phenyl}hex-5-enoic acid, Estrogen receptor, Nuclear receptor coactivator 2 | | 著者 | Nwachukwu, J.C, Srinivasan, S, Bruno, N.E, Nowak, J, Kojetin, D.J, Elemento, O, Katzenellenbogen, J.A, Nettles, K.W. | | 登録日 | 2016-10-13 | | 公開日 | 2017-01-18 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Systems Structural Biology Analysis of Ligand Effects on ER alpha Predicts Cellular Response to Environmental Estrogens and Anti-hormone Therapies.
Cell Chem Biol, 24, 2017
|
|
5TN4
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in Complex with the ACD-ring estrogen, (S)-5-(4-hydroxy-3,5-dimethylphenyl)-2,3-dihydro-1H-inden-1-ol | | 分子名称: | (1S)-5-(4-hydroxy-3,5-dimethylphenyl)-2,3-dihydro-1H-inden-1-ol, Estrogen receptor, Nuclear receptor coactivator 2 | | 著者 | Nwachukwu, J.C, Erumbi, R, Srinivasan, S, Bruno, N.E, Nowak, J, Izard, T, Kojetin, D.J, Elemento, O, Katzenellenbogen, J.A, Nettles, K.W. | | 登録日 | 2016-10-13 | | 公開日 | 2017-01-18 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.857 Å) | | 主引用文献 | Systems Structural Biology Analysis of Ligand Effects on ER alpha Predicts Cellular Response to Environmental Estrogens and Anti-hormone Therapies.
Cell Chem Biol, 24, 2017
|
|
5TNM
 
 | |
3DRA
 
 | | Candida albicans protein geranylgeranyltransferase-I complexed with GGPP | | 分子名称: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GERANYLGERANYL DIPHOSPHATE, Geranylgeranyltransferase type I beta subunit, ... | | 著者 | Hast, M.A, Beese, L.S. | | 登録日 | 2008-07-10 | | 公開日 | 2008-09-30 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structure of protein geranylgeranyltransferase-I from the human pathogen Candida albicans complexed with a lipid substrate.
J.Biol.Chem., 283, 2008
|
|
3QQJ
 
 | | CDK2 in complex with inhibitor L2 | | 分子名称: | 1,2-ETHANEDIOL, 2-(4,6-diamino-1,3,5-triazin-2-yl)phenol, Cyclin-dependent kinase 2, ... | | 著者 | Betzi, S, Alam, R, Han, H, Becker, A, Schonbrunn, E. | | 登録日 | 2011-02-15 | | 公開日 | 2012-08-08 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structure-guided optimization of novel CDK2 inhibitors discovered by high-throughput screening
To be Published
|
|
5BN2
 
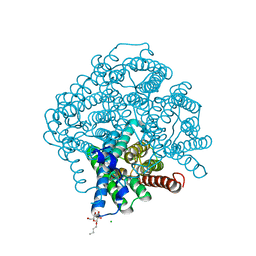 | | Room Temperature Structure of Pichia pastoris aquaporin at 1.3 A | | 分子名称: | AQY1 protein, CALCIUM ION, CHLORIDE ION, ... | | 著者 | Fischer, G, Kosinska Eriksson, U, Hedfalk, K, Neutze, R. | | 登録日 | 2015-05-25 | | 公開日 | 2016-06-29 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | Room Temperature Structure of Pichia pastoris aquaporin at 1.3 A
To Be Published
|
|
5TLB
 
 | | Scabin toxin from Streptomyces scabies in complex with NADH | | 分子名称: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Scabin | | 著者 | Lyons, B, Dutta, D, Ravulapalli, R, Merrill, A.R. | | 登録日 | 2016-10-10 | | 公開日 | 2017-10-11 | | 最終更新日 | 2024-11-13 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Characterization of the catalytic signature of Scabin toxin, a DNA-targeting ADP-ribosyltransferase.
Biochem. J., 475, 2018
|
|
5TM8
 
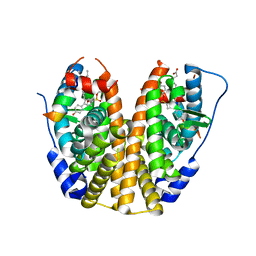 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in Complex with the OBHS-ASC compound, 7-(4-((1R,4S,6R)-6-((4-bromophenoxy)sulfonyl)-3-(4-hydroxyphenyl)-7-oxabicyclo[2.2.1]hept-2-en-2-yl)phenoxy)heptanoic acid | | 分子名称: | 7-{4-[(1S,4S,6R)-6-[(4-bromophenoxy)sulfonyl]-3-(4-hydroxyphenyl)-7-oxabicyclo[2.2.1]hept-2-en-2-yl]phenoxy}heptanoic acid, Estrogen receptor, Nuclear receptor coactivator 2 | | 著者 | Nwachukwu, J.C, Erumbi, R, Srinivasan, S, Bruno, N.E, Nowak, J, Izard, T, Kojetin, D.J, Elemento, O, Katzenellenbogen, J.A, Nettles, K.W. | | 登録日 | 2016-10-12 | | 公開日 | 2017-01-18 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.99 Å) | | 主引用文献 | Systems Structural Biology Analysis of Ligand Effects on ER alpha Predicts Cellular Response to Environmental Estrogens and Anti-hormone Therapies.
Cell Chem Biol, 24, 2017
|
|
7JWU
 
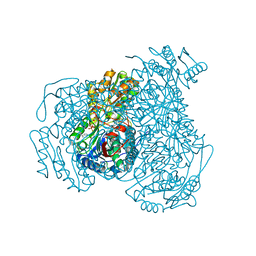 | | Crystal structure of human ALDH1A1 bound to compound (R)-28 | | 分子名称: | 1-methyl-5-phenyl-6-{[(1R)-1-(pyridin-2-yl)ethyl]sulfanyl}-1,5-dihydro-4H-pyrazolo[3,4-d]pyrimidin-4-one, CHLORIDE ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | 著者 | Hurley, T.D, Buchman, C. | | 登録日 | 2020-08-26 | | 公開日 | 2020-12-30 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Development of 2,5-dihydro-4H-pyrazolo[3,4-d]pyrimidin-4-one inhibitors of aldehyde dehydrogenase 1A (ALDH1A) as potential adjuncts to ovarian cancer chemotherapy.
Eur.J.Med.Chem., 211, 2020
|
|
1W4L
 
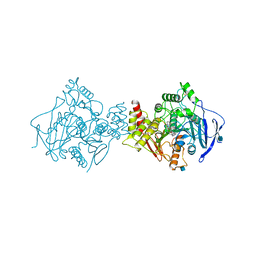 | | Complex of TcAChE with bis-acting galanthamine derivative | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETYLCHOLINESTERASE, ... | | 著者 | Greenblatt, H.M, Guillou, C, Guenard, D, Badet, B, Thal, C, Silman, I, Sussman, J.L. | | 登録日 | 2004-07-25 | | 公開日 | 2004-11-25 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (2.16 Å) | | 主引用文献 | The complex of a bivalent derivative of galanthamine with torpedo acetylcholinesterase displays drastic deformation of the active-site gorge: implications for structure-based drug design.
J.Am.Chem.Soc., 126, 2004
|
|
2WE5
 
 | | Carbamate kinase from Enterococcus faecalis bound to MgADP | | 分子名称: | ACETATE ION, ADENOSINE-5'-DIPHOSPHATE, CARBAMATE KINASE 1, ... | | 著者 | Ramon-Maiques, S, Marina, A, Rubio, V. | | 登録日 | 2009-03-27 | | 公開日 | 2010-03-16 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.39 Å) | | 主引用文献 | Substrate Binding and Catalysis in Carbamate Kinase Ascertained by Crystallographic and Site- Directed Mutagenesis Studies. Movements and Significance of a Unique Globular Subdomain of This Key Enzyme for Fermentative ATP Production in Bacteria.
J.Mol.Biol., 397, 2010
|
|
2W6M
 
 | |
5YAD
 
 | |
3JYA
 
 | | Discovery of 3H-benzo[4,5]thieno[3,2-d]pyrimidin-4-ones as Potent, Highly Selective and Orally Bioavailable Pim Kinases Inhibitors | | 分子名称: | 6,9-dichloro[1]benzothieno[3,2-d]pyrimidin-4(3H)-one, Proto-oncogene serine/threonine-protein kinase Pim-1 | | 著者 | Stoll, V.S. | | 登録日 | 2009-09-21 | | 公開日 | 2009-11-10 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Discovery of 3H-benzo[4,5]thieno[3,2-d]pyrimidin-4-ones as Potent, Highly Selective and Orally Bioavailable Pim Kinases Inhibitors
TO BE PUBLISHED
|
|
7G1E
 
 | | Crystal Structure of human FABP4 in complex with 3-[(2-phenylphenoxy)methyl]benzoic acid, i.e. SMILES c1cc(c(cc1)OCc1cccc(c1)C(=O)O)c1ccccc1 with IC50=0.915 microM | | 分子名称: | 3-{[([1,1'-biphenyl]-2-yl)oxy]methyl}benzoic acid, Fatty acid-binding protein, adipocyte, ... | | 著者 | Ehler, A, Benz, J, Obst, U, Gillespie, P, Rudolph, M.G. | | 登録日 | 2023-04-27 | | 公開日 | 2023-06-14 | | 最終更新日 | 2025-08-13 | | 実験手法 | X-RAY DIFFRACTION (1.21 Å) | | 主引用文献 | A high-resolution data set of fatty acid-binding protein structures. III. Unexpectedly high occurrence of wrong ligands.
Acta Crystallogr D Struct Biol, 81, 2025
|
|
5QCU
 
 | | Crystal structure of BACE complex with BMC022 | | 分子名称: | (2R,4S)-N-butyl-4-[(5S,8S,10R)-5,10-dimethyl-3,3,6-trioxo-3lambda~6~-thia-7-azabicyclo[11.3.1]heptadeca-1(17),13,15-trien-8-yl]-4-hydroxy-2-methylbutanamide, Beta-secretase 1 | | 著者 | Rondeau, J.M, Shao, C, Yang, H, Burley, S.K. | | 登録日 | 2017-12-01 | | 公開日 | 2020-06-03 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (1.951 Å) | | 主引用文献 | D3R grand challenge 4: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J.Comput.Aided Mol.Des., 34, 2020
|
|
4PGN
 
 | | CRYSTAL STRUCTURE OF A TRAP PERIPLASMIC SOLUTE BINDING PROTEIN FROM DESULFOVIBRIO ALASKENSIS G20 (Dde_0634, TARGET EFI-510120) WITH BOUND INDOLE PYRUVATE | | 分子名称: | 1,2-ETHANEDIOL, 3-(1H-INDOL-3-YL)-2-OXOPROPANOIC ACID, Extracellular solute-binding protein, ... | | 著者 | Vetting, M.W, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Stead, M, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | 登録日 | 2014-05-02 | | 公開日 | 2014-05-28 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
5QD6
 
 | | Crystal structure of BACE complex with BMC004 | | 分子名称: | (3S,14R,16S)-16-[1,1-dihydroxy-2-({[3-(propan-2-yl)phenyl]methyl}amino)ethyl]-3,4,14-trimethyl-1,4-diazacyclohexadecane-2,5-dione, Beta-secretase 1 | | 著者 | Rondeau, J.M, Shao, C, Yang, H, Burley, S.K. | | 登録日 | 2017-12-01 | | 公開日 | 2020-06-03 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (2.51 Å) | | 主引用文献 | D3R grand challenge 4: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J.Comput.Aided Mol.Des., 34, 2020
|
|
