6MSH
 
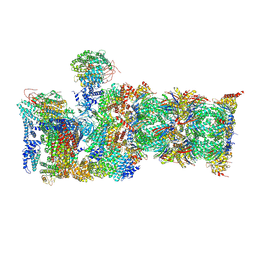 | |
6NYV
 
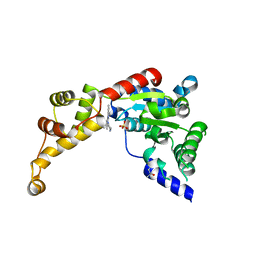 | | Structure of spastin AAA domain in complex with a quinazoline-based inhibitor | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, N-(3-tert-butyl-1H-pyrazol-5-yl)-2-[(2R)-2-methylpiperazin-1-yl]quinazolin-4-amine, SULFATE ION, ... | | 著者 | Pisa, R, Cupido, T, Kapoor, T.M. | | 登録日 | 2019-02-12 | | 公開日 | 2019-03-27 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.425 Å) | | 主引用文献 | Designing Allele-Specific Inhibitors of Spastin, a Microtubule-Severing AAA Protein.
J. Am. Chem. Soc., 141, 2019
|
|
6NYY
 
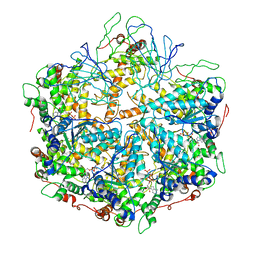 | | human m-AAA protease AFG3L2, substrate-bound | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, AFG3-like protein 2, MAGNESIUM ION, ... | | 著者 | Lander, G.C, Puchades, C. | | 登録日 | 2019-02-12 | | 公開日 | 2019-05-22 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Unique Structural Features of the Mitochondrial AAA+ Protease AFG3L2 Reveal the Molecular Basis for Activity in Health and Disease.
Mol.Cell, 75, 2019
|
|
6NDY
 
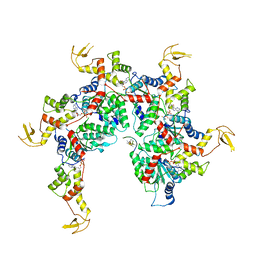 | | Vps4 with Cyclic Peptide Bound in the Central Pore | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, Designed Cyclic Peptide, ... | | 著者 | Han, H, Fulcher, J.M, Dandey, V.P, Sundquist, W.I, Kay, M.S, Shen, P, Hill, C.P. | | 登録日 | 2018-12-14 | | 公開日 | 2019-08-21 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Structure of Vps4 with circular peptides and implications for translocation of two polypeptide chains by AAA+ ATPases.
Elife, 8, 2019
|
|
6MSD
 
 | |
6N8T
 
 | | Hsp104DWB closed conformation | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, Heat shock protein 104 | | 著者 | Lee, S, Rho, S.H, Lee, J, Sung, N, Liu, J, Tsai, F.T.F. | | 登録日 | 2018-11-30 | | 公開日 | 2019-01-02 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (7.7 Å) | | 主引用文献 | Cryo-EM Structures of the Hsp104 Protein Disaggregase Captured in the ATP Conformation.
Cell Rep, 26, 2019
|
|
6MSK
 
 | |
6MDP
 
 | | The D1 and D2 domain rings of NSF engaging the SNAP-25 N-terminus within the 20S supercomplex (focused refinement on D1/D2 rings, class 2) | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Synaptosomal-associated protein 25, ... | | 著者 | White, K.I, Zhao, M, Brunger, A.T. | | 登録日 | 2018-09-04 | | 公開日 | 2018-09-19 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Structural principles of SNARE complex recognition by the AAA+ protein NSF.
Elife, 7, 2018
|
|
6NYW
 
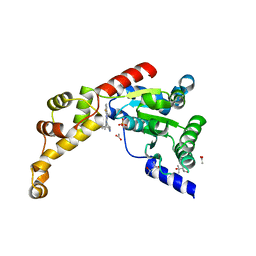 | | Structure of spastin AAA domain N527C mutant in complex with 8-fluoroquinazoline-based inhibitor | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, N-(5-tert-butyl-1H-pyrazol-3-yl)-8-fluoro-2-[(2R)-2-methylpiperazin-1-yl]quinazolin-4-amine, ... | | 著者 | Pisa, R, Cupido, T, Kapoor, T.M. | | 登録日 | 2019-02-12 | | 公開日 | 2019-03-27 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.189 Å) | | 主引用文献 | Designing Allele-Specific Inhibitors of Spastin, a Microtubule-Severing AAA Protein.
J. Am. Chem. Soc., 141, 2019
|
|
6MSG
 
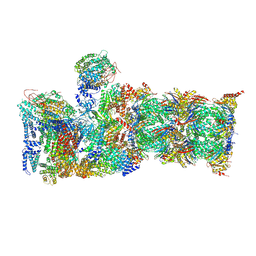 | |
6OAA
 
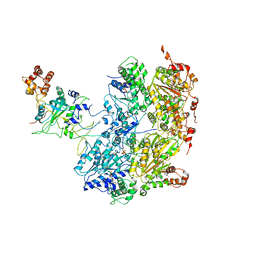 | | Cdc48-Npl4 complex processing poly-ubiquitinated substrate in the presence of ADP-BeFx, state 1 | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, Cell division control protein 48, ... | | 著者 | Twomey, E.C, Ji, Z, Wales, T.E, Bodnar, N.O, Engen, J.R, Rapoport, T.A. | | 登録日 | 2019-03-15 | | 公開日 | 2019-07-03 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (4.1 Å) | | 主引用文献 | Substrate processing by the Cdc48 ATPase complex is initiated by ubiquitin unfolding.
Science, 365, 2019
|
|
6OA9
 
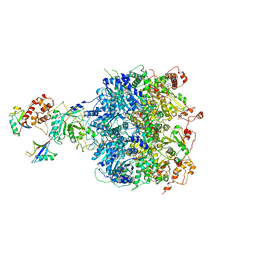 | | Cdc48-Npl4 complex processing poly-ubiquitinated substrate in the presence of ATP | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Cell division control protein 48, ... | | 著者 | Twomey, E.C, Ji, Z, Wales, T.E, Bodnar, N.O, Engen, J.R, Rapoport, T.A. | | 登録日 | 2019-03-15 | | 公開日 | 2019-07-03 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Substrate processing by the Cdc48 ATPase complex is initiated by ubiquitin unfolding.
Science, 365, 2019
|
|
6OG2
 
 | | Focus classification structure of the hyperactive ClpB mutant K476C, bound to casein, post-state | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Hyperactive disaggregase ClpB | | 著者 | Rizo, A.R, Lin, J.-B, Gates, S.N, Tse, E, Bart, S.M, Castellano, L.M, Dimaio, F, Shorter, J, Southworth, D.R. | | 登録日 | 2019-04-01 | | 公開日 | 2019-06-12 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (4.1 Å) | | 主引用文献 | Structural basis for substrate gripping and translocation by the ClpB AAA+ disaggregase.
Nat Commun, 10, 2019
|
|
6OMB
 
 | | Cdc48 Hexamer (Subunits A to E) with substrate bound to the central pore | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, Cell division control protein 48, ... | | 著者 | Cooney, I, Han, H, Stewart, M, Carson, R.H, Hansen, D, Price, J.C, Hill, C.P, Shen, P.S. | | 登録日 | 2019-04-18 | | 公開日 | 2019-07-17 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Structure of the Cdc48 segregase in the act of unfolding an authentic substrate.
Science, 365, 2019
|
|
6OAX
 
 | | Structure of the hyperactive ClpB mutant K476C, bound to casein, pre-state | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Alpha-S1-casein, Hyperactive disaggregase ClpB, ... | | 著者 | Rizo, A.R, Lin, J.-B, Gates, S.N, Tse, E, Bart, S.M, Castellano, L.M, Dimaio, F, Shorter, J, Southworth, D.R. | | 登録日 | 2019-03-18 | | 公開日 | 2019-06-12 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Structural basis for substrate gripping and translocation by the ClpB AAA+ disaggregase.
Nat Commun, 10, 2019
|
|
5TXV
 
 | | HslU P21 cell with 4 hexamers | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent protease ATPase subunit HslU | | 著者 | Grant, R.A, Chen, J, Glynn, S.E, Sauer, R.T. | | 登録日 | 2016-11-17 | | 公開日 | 2017-03-01 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (7.086 Å) | | 主引用文献 | Covalently linked HslU hexamers support a probabilistic mechanism that links ATP hydrolysis to protein unfolding and translocation.
J. Biol. Chem., 292, 2017
|
|
5UBV
 
 | |
5UJM
 
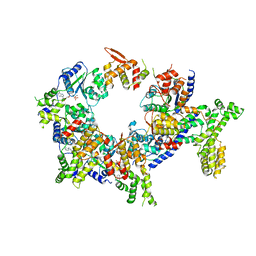 | | Structure of the active form of human Origin Recognition Complex and its ATPase motor module | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Origin recognition complex subunit 1, ... | | 著者 | Tocilj, A, On, K, Yuan, Z, Sun, J, Elkayam, E, Li, H, Stillman, B, Joshua-Tor, L. | | 登録日 | 2017-01-18 | | 公開日 | 2017-02-08 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (18 Å) | | 主引用文献 | Structure of the active form of human Origin Recognition Complex and its ATPase motor module.
Elife, 6, 2017
|
|
5UIE
 
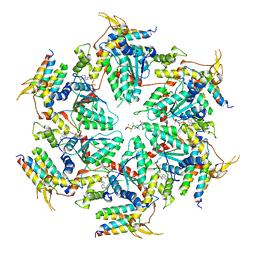 | | Vps4-Vta1 complex | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, DOA4-independent degradation protein 4, ... | | 著者 | Monroe, N, Shen, P, Han, H, Sundquist, W.I, Hill, C.P. | | 登録日 | 2017-01-13 | | 公開日 | 2017-04-12 | | 最終更新日 | 2020-01-01 | | 実験手法 | ELECTRON MICROSCOPY (5.7 Å) | | 主引用文献 | Structural basis of protein translocation by the Vps4-Vta1 AAA ATPase.
Elife, 6, 2017
|
|
5T0I
 
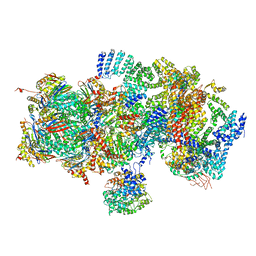 | | Structural basis for dynamic regulation of the human 26S proteasome | | 分子名称: | 26S protease regulatory subunit 10B, 26S protease regulatory subunit 4, 26S protease regulatory subunit 6A, ... | | 著者 | Chen, S, Wu, J, Lu, Y, Ma, Y.B, Lee, B.H, Yu, Z, Ouyang, Q, Finley, D, Kirschner, M.W, Mao, Y. | | 登録日 | 2016-08-16 | | 公開日 | 2016-10-19 | | 最終更新日 | 2016-11-30 | | 実験手法 | ELECTRON MICROSCOPY (8 Å) | | 主引用文献 | Structural basis for dynamic regulation of the human 26S proteasome.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5T0C
 
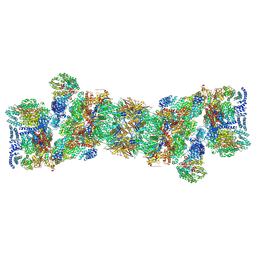 | | Structural basis for dynamic regulation of the human 26S proteasome | | 分子名称: | 26S protease regulatory subunit 10B, 26S protease regulatory subunit 4, 26S protease regulatory subunit 6A, ... | | 著者 | Chen, S, Wu, J, Lu, Y, Ma, Y.B, Lee, B.H, Yu, Z, Ouyang, Q, Finley, D, Kirschner, M.W, Mao, Y. | | 登録日 | 2016-08-15 | | 公開日 | 2016-10-19 | | 最終更新日 | 2018-07-18 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Structural basis for dynamic regulation of the human 26S proteasome.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5VC7
 
 | | VCP like ATPase from T. acidophilum (VAT) - conformation 1 | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, VCP-like ATPase | | 著者 | Ripstein, Z.A, Huang, R, Augustyniak, R, Kay, L.E, Rubinstein, J.L. | | 登録日 | 2017-03-31 | | 公開日 | 2017-04-26 | | 最終更新日 | 2020-01-15 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Structure of a AAA+ unfoldase in the process of unfolding substrate.
Elife, 6, 2017
|
|
5V8F
 
 | | Structural basis of MCM2-7 replicative helicase loading by ORC-Cdc6 and Cdt1 | | 分子名称: | Cell division control protein 6, Cell division cycle protein CDT1, DNA (39-MER), ... | | 著者 | Yuan, Z, Riera, A, Bai, L, Sun, J, Spanos, C, Chen, Z.A, Barbon, M, Rappsilber, J, Stillman, B, Speck, C, Li, H. | | 登録日 | 2017-03-21 | | 公開日 | 2017-05-10 | | 最終更新日 | 2024-10-09 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Structural basis of Mcm2-7 replicative helicase loading by ORC-Cdc6 and Cdt1.
Nat. Struct. Mol. Biol., 24, 2017
|
|
5VFP
 
 | | Nucleotide-driven Triple-state Remodeling of the AAA-ATPase Channel in the Activated Human 26S Proteasome | | 分子名称: | 26S proteasome non-ATPase regulatory subunit 1, 26S proteasome non-ATPase regulatory subunit 11, 26S proteasome non-ATPase regulatory subunit 12, ... | | 著者 | Zhu, Y, Wang, W.L, Yu, D, Ouyang, Q, Lu, Y, Mao, Y. | | 登録日 | 2017-04-09 | | 公開日 | 2018-07-18 | | 実験手法 | ELECTRON MICROSCOPY (4.2 Å) | | 主引用文献 | Structural mechanism for nucleotide-driven remodeling of the AAA-ATPase unfoldase in the activated human 26S proteasome.
Nat Commun, 9, 2018
|
|
5VHR
 
 | | Conformational Landscape of the p28-Bound Human Proteasome Regulatory Particle | | 分子名称: | 26S proteasome non-ATPase regulatory subunit 10, 26S proteasome non-ATPase regulatory subunit 2, 26S proteasome regulatory subunit 10B, ... | | 著者 | Lu, Y, Wu, J, Dong, Y, Chen, S, Sun, S, Ma, Y.B, Ouyang, Q, Finley, D, Kirschner, M.W, Mao, Y. | | 登録日 | 2017-04-13 | | 公開日 | 2017-08-23 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (7.7 Å) | | 主引用文献 | Conformational Landscape of the p28-Bound Human Proteasome Regulatory Particle.
Mol. Cell, 67, 2017
|
|
