5SRF
 
 | |
5FHB
 
 | |
6E9L
 
 | |
4Q0M
 
 | | Crystal structure of Pyrococcus furiosus L-asparaginase | | 分子名称: | (4S)-2-METHYL-1,4,5,6-TETRAHYDROPYRIMIDINE-4-CARBOXYLIC ACID, GLYCEROL, L-asparaginase, ... | | 著者 | Sharma, P, Tomar, R, Singh, S, Yadav, S.P.S, Ashish, Kundu, B. | | 登録日 | 2014-04-02 | | 公開日 | 2014-12-10 | | 最終更新日 | 2024-11-13 | | 実験手法 | X-RAY DIFFRACTION (2.226 Å) | | 主引用文献 | Structural and functional insights into an archaeal L-asparaginase obtained through the linker-less assembly of constituent domains.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
6KTN
 
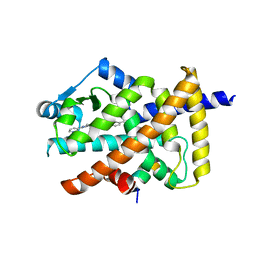 | | Human PPARgamma ligand-binding domain R288A mutant in complex with imatinib | | 分子名称: | 16-mer peptide from Nuclear receptor coactivator 1, 4-(4-METHYL-PIPERAZIN-1-YLMETHYL)-N-[4-METHYL-3-(4-PYRIDIN-3-YL-PYRIMIDIN-2-YLAMINO)-PHENYL]-BENZAMIDE, Peroxisome proliferator-activated receptor gamma | | 著者 | Jang, J.Y, Han, B.W. | | 登録日 | 2019-08-28 | | 公開日 | 2020-02-05 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.752 Å) | | 主引用文献 | Structural Basis for the Regulation of PPAR gamma Activity by Imatinib.
Molecules, 24, 2019
|
|
4Q1H
 
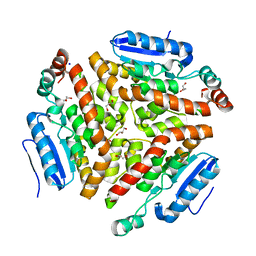 | | Structure and mechanism of a dehydratase/decarboxylase enzyme couple involved in polyketide beta-branching | | 分子名称: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, Polyketide biosynthesis enoyl-CoA isomerase PksI, ... | | 著者 | Nair, A.V, Race, P.R, Till, M. | | 登録日 | 2014-04-03 | | 公開日 | 2015-05-06 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (1.93 Å) | | 主引用文献 | Structure and mechanism of a dehydratase/decarboxylase enzyme couple involved in polyketide beta-methyl branch incorporation.
Sci Rep, 10, 2020
|
|
3IKJ
 
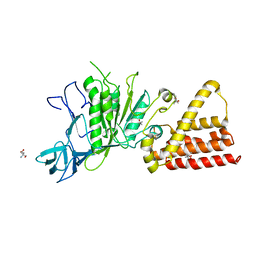 | | Structural characterization for the nucleotide binding ability of subunit A mutant S238A of the A1AO ATP synthase | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, V-type ATP synthase alpha chain | | 著者 | Kumar, A, Manimekali, M.S.S, Balakrishna, A.M, Jeyakanthan, J, Gruber, G. | | 登録日 | 2009-08-06 | | 公開日 | 2010-01-12 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Nucleotide binding states of subunit A of the A-ATP synthase and the implication of P-loop switch in evolution.
J.Mol.Biol., 396, 2010
|
|
2VX7
 
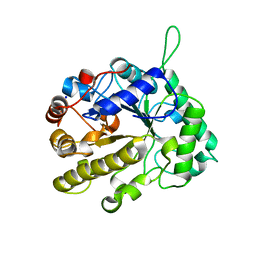 | | CELLVIBRIO JAPONICUS MANNANASE CJMAN26C MANNOBIOSE-BOUND FORM | | 分子名称: | CELLVIBRIO JAPONICUS MANNANASE CJMAN26C, SODIUM ION, beta-D-mannopyranose-(1-4)-beta-D-mannopyranose | | 著者 | Cartmell, A, Topakas, E, Ducros, V.M.-A, Suits, M.D.L, Davies, G.J, Gilbert, H.J. | | 登録日 | 2008-07-01 | | 公開日 | 2008-09-16 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | The Cellvibrio Japonicus Mannanase Cjman26C Displays a Unique Exo-Mode of Action that is Conferred by Subtle Changes to the Distal Region of the Active Site.
J.Biol.Chem., 283, 2008
|
|
6AJH
 
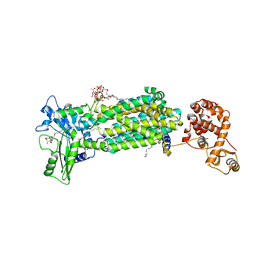 | | Crystal structure of mycolic acid transporter MmpL3 from Mycobacterium smegmatis complexed with AU1235 | | 分子名称: | (CARBAMOYLMETHYL-CARBOXYMETHYL-AMINO)-ACETIC ACID, 1-(2-adamantyl)-3-[2,3,4-tris(fluoranyl)phenyl]urea, Drug exporters of the RND superfamily-like protein,Endolysin, ... | | 著者 | Zhang, B, Li, J, Yang, X.L, Wu, L.J, Yang, H.T, Rao, Z.H. | | 登録日 | 2018-08-27 | | 公開日 | 2018-12-26 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.818 Å) | | 主引用文献 | Crystal Structures of Membrane Transporter MmpL3, an Anti-TB Drug Target.
Cell, 176, 2019
|
|
5BSX
 
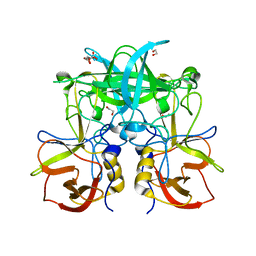 | |
5SQD
 
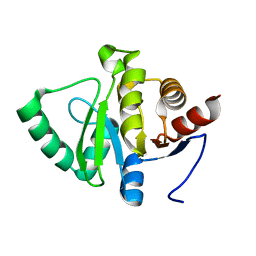 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with Z5010894382 - (R,R) and (S,S) isomers | | 分子名称: | (1R,2R)-4-hydroxy-1-{[2-(hydroxymethyl)-1H-benzimidazole-5-carbonyl]amino}-2,3-dihydro-1H-indene-2-carboxylic acid, (1S,2S)-4-hydroxy-1-{[2-(hydroxymethyl)-1H-benzimidazole-5-carbonyl]amino}-2,3-dihydro-1H-indene-2-carboxylic acid, Non-structural protein 3 | | 著者 | Correy, G.J, Fraser, J.S. | | 登録日 | 2022-06-09 | | 公開日 | 2022-07-06 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.05 Å) | | 主引用文献 | Iterative computational design and crystallographic screening identifies potent inhibitors targeting the Nsp3 macrodomain of SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
4XNK
 
 | | X-ray structure of AlgE1 | | 分子名称: | (2S)-2,3-DIHYDROXYPROPYL(7Z)-PENTADEC-7-ENOATE, 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, Alginate production protein AlgE, ... | | 著者 | Ma, P, Huang, C.Y, Olieric, V, Diederichs, K, Wang, M, Caffrey, M. | | 登録日 | 2015-01-15 | | 公開日 | 2015-06-03 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | In meso in situ serial X-ray crystallography of soluble and membrane proteins.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
5SQE
 
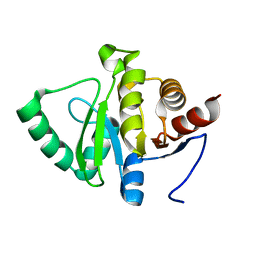 | |
5X35
 
 | |
8SA2
 
 | | Adenosylcobalamin-bound riboswitch dimer, form 1 | | 分子名称: | Adenosylcobalamin, adenosylcobalamin riboswitch form 1 | | 著者 | Ding, J, Deme, J.C, Stagno, J.R, Yu, P, Lea, S.M, Wang, Y.X. | | 登録日 | 2023-03-31 | | 公開日 | 2023-07-26 | | 最終更新日 | 2025-06-04 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Capturing heterogeneous conformers of cobalamin riboswitch by cryo-EM.
Nucleic Acids Res., 51, 2023
|
|
4XQJ
 
 | |
5SQM
 
 | |
4QBW
 
 | | The second sphere residue T263 is important for function and activity of PTP1B through modulating WPD loop | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Tyrosine-protein phosphatase non-receptor type 1 | | 著者 | Xiao, P, Wang, X, Wang, H.M, Fu, X.L, Cui, F.A, Yu, X, Bi, W.X, Sun, J.P. | | 登録日 | 2014-05-08 | | 公開日 | 2015-02-11 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.912 Å) | | 主引用文献 | The second-sphere residue T263 is important for the function and catalytic activity of PTP1B via interaction with the WPD-loop
Int.J.Biochem.Cell Biol., 57, 2014
|
|
5SQL
 
 | |
7EM1
 
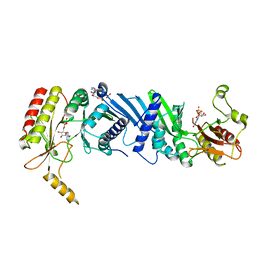 | | Crystal structure of the PI5P4Kbeta-ITP complex | | 分子名称: | INOSINE-5'-DIPHOSPHATE, Phosphatidylinositol 5-phosphate 4-kinase type-2 beta, [[(2~{R},3~{S},4~{R},5~{R})-3,4-bis(oxidanyl)-5-(6-oxidanylidene-1~{H}-purin-9-yl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] phosphono hydrogen phosphate | | 著者 | Senda, M, Senda, T. | | 登録日 | 2021-04-13 | | 公開日 | 2022-03-30 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.65 Å) | | 主引用文献 | The GTP responsiveness of PI5P4K beta evolved from a compromised trade-off between activity and specificity.
Structure, 30, 2022
|
|
4QDJ
 
 | |
6EI2
 
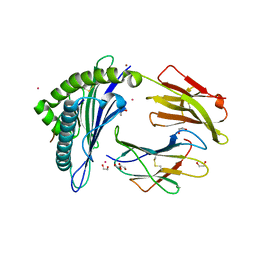 | | Crystal Structure of HLA-A68 presenting a C-terminally extended peptide | | 分子名称: | 1,2-ETHANEDIOL, Beta-2-microglobulin, CADMIUM ION, ... | | 著者 | Picaud, S, Guillaume, P, Pike, A.C.W, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Gfeller, D, Filippakopoulos, P. | | 登録日 | 2017-09-16 | | 公開日 | 2017-10-11 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (1.61 Å) | | 主引用文献 | Crystal Structure of HLA-A68 presenting a C-terminally extended peptide
To Be Published
|
|
7EI0
 
 | |
5BN3
 
 | | Structure of a unique ATP synthase NeqA-NeqB in complex with ADP from Nanoarcheaum equitans | | 分子名称: | 1,4-DIETHYLENE DIOXIDE, ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, ... | | 著者 | Mohanty, S, Jobichen, C, Chichili, V.P.R, Sivaraman, J. | | 登録日 | 2015-05-25 | | 公開日 | 2015-09-16 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural Basis for a Unique ATP Synthase Core Complex from Nanoarcheaum equitans
J.Biol.Chem., 290, 2015
|
|
5SFY
 
 | | CRYSTAL STRUCTURE OF HUMAN PHOSPHODIESTERASE 10 IN COMPLEX WITH n2c(c1nc(nn1c(c2)C)C=Cc3nc(nn3C)c4c[nH]nc4)C, micromolar IC50=0.22176 | | 分子名称: | (4S)-5,8-dimethyl-2-{(E)-2-[1-methyl-3-(1H-pyrazol-4-yl)-1H-1,2,4-triazol-5-yl]ethenyl}[1,2,4]triazolo[1,5-a]pyrazine, MAGNESIUM ION, ZINC ION, ... | | 著者 | Joseph, C, Flohr, A, Benz, J, Schlatter, D, Rudolph, M.G. | | 登録日 | 2022-01-21 | | 公開日 | 2022-10-12 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (2.08 Å) | | 主引用文献 | A high quality, industrial data set for binding affinity prediction: performance comparison in different early drug discovery scenarios.
J.Comput.Aided Mol.Des., 36, 2022
|
|
