1MR4
 
 | | Solution Structure of NaD1 from Nicotiana alata | | 分子名称: | Nicotiana alata plant defensin 1 (NaD1) | | 著者 | Lay, F.T, Schirra, H.J, Scanlon, M.J, Anderson, M.A, Craik, D.J. | | 登録日 | 2002-09-18 | | 公開日 | 2003-09-18 | | 最終更新日 | 2022-02-23 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The Three-dimensional Solution Structure of NaD1, a New Floral Defensin from Nicotiana alata and its Application to a Homology Model of the Crop Defense Protein alfAFP
J.MOL.BIOL., 325, 2003
|
|
1Q8X
 
 | | NMR structure of human cofilin | | 分子名称: | Cofilin, non-muscle isoform | | 著者 | Pope, B.J, Zierler-Gould, K.M, Kuhne, R, Weeds, A.G, Ball, L.J. | | 登録日 | 2003-08-22 | | 公開日 | 2004-07-06 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The solution structure of human cofilin: rationalizing actin binding and pH sensitivity
J.Biol.Chem., 279, 2004
|
|
6HYK
 
 | | NMR solution structure of the C/D box snoRNA U14 | | 分子名称: | RNA (31-MER) | | 著者 | Chagot, M.E, Quinternet, M, Rothe, B, Charpentier, B, Coutant, J, Manival, X, Lebars, I. | | 登録日 | 2018-10-22 | | 公開日 | 2019-04-24 | | 最終更新日 | 2024-06-19 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The yeast C/D box snoRNA U14 adopts a "weak" K-turn like conformation recognized by the Snu13 core protein in solution.
Biochimie, 164, 2019
|
|
6IY5
 
 | | NMR solution structures of 5'-ATTCTATTCT-3 | | 分子名称: | DNA (5'-D(*AP*TP*TP*CP*TP*AP*TP*TP*CP*T)-3'), SODIUM ION | | 著者 | Lam, S.L, Guo, P. | | 登録日 | 2018-12-13 | | 公開日 | 2020-06-10 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Minidumbbell structures formed by ATTCT pentanucleotide repeats in spinocerebellar ataxia type 10.
Nucleic Acids Res., 48, 2020
|
|
1U6P
 
 | |
6GS5
 
 | |
1RY4
 
 | |
1JJD
 
 | |
6TV5
 
 | |
5X29
 
 | |
1L3E
 
 | | NMR Structures of the HIF-1alpha CTAD/p300 CH1 Complex | | 分子名称: | ZINC ION, hypoxia inducible factor-1 alpha subunit, p300 protein | | 著者 | Freedman, S.J, Sun, Z.J, Poy, F, Kung, A.L, Livingston, D.M, Wagner, G, Eck, M.J. | | 登録日 | 2002-02-26 | | 公開日 | 2002-04-24 | | 最終更新日 | 2022-02-23 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural basis for recruitment of CBP/p300 by hypoxia-inducible factor-1 alpha.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
4BS2
 
 | | NMR structure of human TDP-43 tandem RRMs in complex with UG-rich RNA | | 分子名称: | 5'-R(*GP*UP*GP*UP*GP*AP*AP*UP*GP*AP*AP*UP)-3', TAR DNA-BINDING PROTEIN 43 | | 著者 | Lukavsky, P.J, Daujotyte, D, Tollervey, J.R, Ule, J, Stuani, C, Buratti, E, Baralle, F.E, Damberger, F.F, Allain, F.H.T. | | 登録日 | 2013-06-06 | | 公開日 | 2013-11-13 | | 最終更新日 | 2024-06-19 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Molecular Basis of Ug-Rich RNA Recognition by the Human Splicing Factor Tdp-43
Nat.Struct.Mol.Biol., 20, 2013
|
|
2Z2G
 
 | | NMR Structure of the IQ-modified Dodecamer CTC[IQ]GGCGCCATC | | 分子名称: | 3-METHYL-3H-IMIDAZO[4,5-F]QUINOLIN-2-AMINE, DNA (5'-D(*DCP*DTP*DCP*DGP*DGP*DCP*DGP*DCP*DCP*DAP*DTP*DC)-3'), DNA (5'-D(*DGP*DAP*DTP*DGP*DGP*DCP*DGP*DCP*DCP*DGP*DAP*DG)-3') | | 著者 | Wang, F, Elmquist, C.E, Stover, J.S, Rizzo, C.J, Stone, M.P. | | 登録日 | 2007-05-22 | | 公開日 | 2007-10-02 | | 最終更新日 | 2023-11-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | DNA sequence modulates the conformation of the food mutagen 2-amino-3-methylimidazo[4,5-f]quinoline in the recognition sequence of the NarI restriction enzyme
Biochemistry, 46, 2007
|
|
2Z2H
 
 | | NMR Structure of the IQ-modified Dodecamer CTCG[IQ]GCGCCATC | | 分子名称: | 3-METHYL-3H-IMIDAZO[4,5-F]QUINOLIN-2-AMINE, DNA (5'-D(*DCP*DTP*DCP*DGP*DGP*DCP*DGP*DCP*DCP*DAP*DTP*DC)-3'), DNA (5'-D(*DGP*DAP*DTP*DGP*DGP*DCP*DGP*DCP*DCP*DGP*DAP*DG)-3') | | 著者 | Wang, F, Elmquist, C.E, Stover, J.S, Rizzo, C.J, Stone, M.P. | | 登録日 | 2007-05-22 | | 公開日 | 2007-10-02 | | 最終更新日 | 2023-11-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | DNA sequence modulates the conformation of the food mutagen 2-amino-3-methylimidazo[4,5-f]quinoline in the recognition sequence of the NarI restriction enzyme
Biochemistry, 46, 2007
|
|
1IEO
 
 | | SOLUTION STRUCTURE OF MRIB-NH2 | | 分子名称: | PROTEIN MRIB-NH2 | | 著者 | Sharpe, I.A, Gehrmann, J, Loughnan, M.L, Thomas, L, Adams, D.A, Atkins, A, Palant, E, Craik, D.J, Adams, D.J, Alewood, P.F, Lewis, R.J. | | 登録日 | 2001-04-10 | | 公開日 | 2002-04-03 | | 最終更新日 | 2022-02-23 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Two new classes of conopeptides inhibit the alpha1-adrenoceptor and noradrenaline transporter.
Nat.Neurosci., 4, 2001
|
|
1CJG
 
 | | NMR STRUCTURE OF LAC REPRESSOR HP62-DNA COMPLEX | | 分子名称: | DNA (5'-D(*GP*AP*AP*TP*TP*GP*TP*GP*AP*GP*CP*GP*CP*TP*CP*AP*CP*AP*AP*TP*TP*C)-3'), PROTEIN (LAC REPRESSOR) | | 著者 | Spronk, C.A.E.M, Bonvin, A.M.J.J, Radha, P.K, Melacini, G, Boelens, R, Kaptein, R. | | 登録日 | 1999-04-14 | | 公開日 | 2000-01-01 | | 最終更新日 | 2023-12-27 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The solution structure of Lac repressor headpiece 62 complexed to a symmetrical lac operator.
Structure Fold.Des., 7, 1999
|
|
6FBL
 
 | |
6AZA
 
 | |
1TBK
 
 | |
1HJ7
 
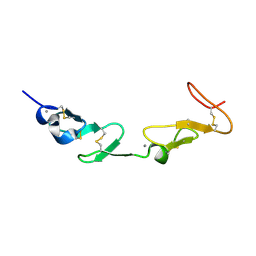 | | NMR study of a pair of LDL receptor Ca2+ binding epidermal growth factor-like domains, 20 structures | | 分子名称: | CALCIUM ION, LDL RECEPTOR | | 著者 | Saha, S, Handford, P.A, Campbell, I.D, Downing, A.K. | | 登録日 | 2001-01-09 | | 公開日 | 2001-07-11 | | 最終更新日 | 2018-02-14 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution Structure of the Ldl Receptor Egf-Ab Pair: A Paradigm for the Assembly of Tandem Calcium Binding Egf Domains
Structure, 9, 2001
|
|
1JSP
 
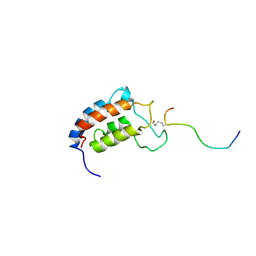 | | NMR Structure of CBP Bromodomain in complex with p53 peptide | | 分子名称: | CREB-BINDING PROTEIN, tumor protein p53 | | 著者 | He, Y, Mujtaba, S, Zeng, L, Yan, S, Zhou, M.-M. | | 登録日 | 2001-08-17 | | 公開日 | 2002-08-17 | | 最終更新日 | 2023-11-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural mechanism of the bromodomain of the coactivator CBP in p53 transcriptional activation.
Mol.Cell, 13, 2004
|
|
1HFN
 
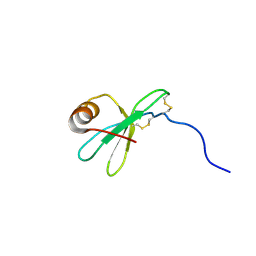 | | NMR solution structures of vMIP-II 1-71 from Kaposi's sarcoma-associated herpesvirus. | | 分子名称: | VIRAL MACROPHAGE INFLAMMATORY PROTEIN-II | | 著者 | Crump, M.P, Elisseeva, E, Gong, J.-H, Clark-Lewis, I, Sykes, B.D. | | 登録日 | 2000-12-07 | | 公開日 | 2001-01-07 | | 最終更新日 | 2011-07-13 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structure/Function of Human Herpesvirus-8 Mip-II (1-71) and the Antagonist N-Terminal Segment (1-10)
FEBS Lett., 489, 2001
|
|
1NGO
 
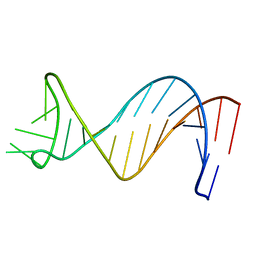 | | NMR Structure of Putative 3' Terminator for B. Anthracis pagA Gene Coding Strand | | 分子名称: | 5'-D(*CP*TP*CP*TP*TP*TP*TP*TP*GP*TP*AP*AP*GP*AP*AP*AP*TP*AP*CP*AP*AP*GP*GP*AP*GP*AP*G)-3' | | 著者 | Shiflett, P.R, Taylor-McCabe, K.J, Michalczyk, R, Silks, L.A, Gupta, G. | | 登録日 | 2002-12-17 | | 公開日 | 2003-06-10 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural Studies on the Hairpins at the 3' Untranslated Region of an Anthrax Toxin Gene
Biochemistry, 42, 2003
|
|
1OQ2
 
 | | NMR structure of hemimethylated GATC site | | 分子名称: | 5'-D(*CP*GP*CP*AP*GP*(6MA)P*TP*CP*TP*CP*GP*C)-3', 5'-D(*GP*CP*GP*AP*GP*AP*TP*CP*TP*GP*CP*G)-3' | | 著者 | Bae, S.-H, Cheong, H.-K, Kang, S, Hwang, D.S, Cheong, C, Choi, B.-S. | | 登録日 | 2003-03-07 | | 公開日 | 2004-04-27 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structure and dynamics of hemimethylated GATC sites: implications for DNA-SeqA recognition
J.Biol.Chem., 278, 2003
|
|
1TTK
 
 | | NMR solution structure of omega-conotoxin MVIIA, a N-type calcium channel blocker | | 分子名称: | Omega-conotoxin MVIIa | | 著者 | Adams, D.J, Smith, A.B, Schroeder, C.I, Yasuda, T, Lewis, R.J. | | 登録日 | 2004-06-22 | | 公開日 | 2004-07-06 | | 最終更新日 | 2022-03-02 | | 実験手法 | SOLUTION NMR | | 主引用文献 | omega-conotoxin CVID inhibits a pharmacologically distinct voltage-sensitive calcium channel associated with transmitter release from preganglionic nerve terminals
J.Biol.Chem., 278, 2003
|
|
