5KWZ
 
 | |
5KX0
 
 | |
5KWP
 
 | |
5KVN
 
 | |
5KX1
 
 | |
5LFN
 
 | |
3Q9N
 
 | | In silico and in vitro co-evolution of a high affinity complementary protein-protein interface | | 分子名称: | CARBAMOYL SARCOSINE, COENZYME A, CoA binding protein, ... | | 著者 | Karanicolas, J, Corn, J.E, Chen, I, Joachimiak, L.A, Dym, O, Chung, S, Albeck, S, Unger, T, Hu, W, Liu, G, Delbecq, S, Montelione, G.T, Spiegel, C, Liu, D, Baker, D, Israel Structural Proteomics Center (ISPC) | | 登録日 | 2011-01-09 | | 公開日 | 2011-04-27 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | A de novo protein binding pair by computational design and directed evolution.
Mol.Cell, 42, 2011
|
|
3QA9
 
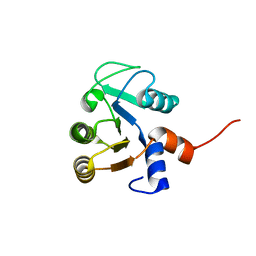 | |
3R3K
 
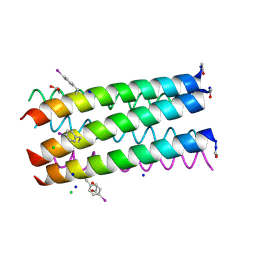 | | Crystal structure of a parallel 6-helix coiled coil | | 分子名称: | 1,2-ETHANEDIOL, CChex-Phi22 helix, CHLORIDE ION, ... | | 著者 | Zaccai, N.R, Chi, B.H.C, Woolfson, D.N, Brady, R.L. | | 登録日 | 2011-03-16 | | 公開日 | 2011-11-16 | | 最終更新日 | 2011-11-30 | | 実験手法 | X-RAY DIFFRACTION (2.2009 Å) | | 主引用文献 | A de novo peptide hexamer with a mutable channel.
Nat.Chem.Biol., 7, 2011
|
|
3R46
 
 | | Crystal structure of a parallel 6-helix coiled coil CC-hex-D24 | | 分子名称: | CHLORIDE ION, GLYCEROL, SODIUM ION, ... | | 著者 | Zaccai, N.R, Chi, B.H.C, Woolfson, D.N, Brady, R.L. | | 登録日 | 2011-03-17 | | 公開日 | 2011-11-16 | | 最終更新日 | 2024-10-09 | | 実験手法 | X-RAY DIFFRACTION (1.751 Å) | | 主引用文献 | A de novo peptide hexamer with a mutable channel.
Nat.Chem.Biol., 7, 2011
|
|
3R4A
 
 | |
3R48
 
 | | Crystal structure of a hetero-hexamer coiled coil | | 分子名称: | GLYCEROL, coiled coil helix W22-L24H, coiled coil helix Y15-L24D | | 著者 | Zaccai, N.R, Chi, B.H.C, Woolfson, D.N, Brady, R.L. | | 登録日 | 2011-03-17 | | 公開日 | 2011-11-16 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.0011 Å) | | 主引用文献 | A de novo peptide hexamer with a mutable channel.
Nat.Chem.Biol., 7, 2011
|
|
3R47
 
 | | Crystal structure of a 6-helix coiled coil CC-hex-H24 | | 分子名称: | BROMIDE ION, coiled coil helix L24H | | 著者 | Zaccai, N.R, Chi, B.H.C, Woolfson, D.N, Brady, R.L. | | 登録日 | 2011-03-17 | | 公開日 | 2011-11-16 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.5002 Å) | | 主引用文献 | A de novo peptide hexamer with a mutable channel.
Nat.Chem.Biol., 7, 2011
|
|
1S48
 
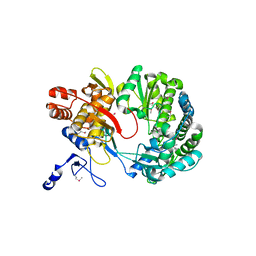 | | Crystal structure of RNA-dependent RNA polymerase construct 1 (residues 71-679) from BVDV | | 分子名称: | RNA-dependent RNA polymerase | | 著者 | Choi, K.H, Groarke, J.M, Young, D.C, Kuhn, R.J, Smith, J.L, Pevear, D.C, Rossmann, M.G. | | 登録日 | 2004-01-15 | | 公開日 | 2004-04-06 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | The structure of the RNA-dependent RNA polymerase from bovine viral diarrhea virus establishes the role of GTP in de novo initiation.
Proc.Natl.Acad.Sci.Usa, 101, 2004
|
|
1S4F
 
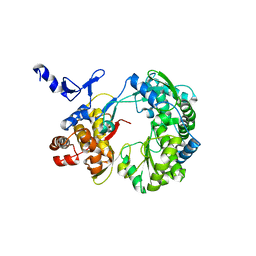 | | Crystal Structure of RNA-dependent RNA polymerase construct 2 from bovine viral diarrhea virus (BVDV) | | 分子名称: | RNA-dependent RNA polymerase | | 著者 | Choi, K.H, Groarke, J.M, Young, D.C, Kuhn, R.J, Smith, J.L, Pevear, D.C, Rossmann, M.G. | | 登録日 | 2004-01-16 | | 公開日 | 2004-04-06 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | The structure of the RNA-dependent RNA polymerase from bovine viral diarrhea virus establishes the role of GTP in de novo initiation.
Proc.Natl.Acad.Sci.Usa, 101, 2004
|
|
1S49
 
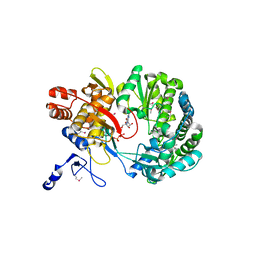 | | Crystal Structure of RNA-dependent RNA polymerase construct 1 (residues 71-679) from bovine viral diarrhea virus complexed with GTP | | 分子名称: | GUANOSINE-5'-TRIPHOSPHATE, RNA-dependent RNA polymerase | | 著者 | Choi, K.H, Groarke, J.M, Young, D.C, Kuhn, R.J, Smith, J.L, Pevear, D.C, Rossmann, M.G. | | 登録日 | 2004-01-15 | | 公開日 | 2004-04-06 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | The structure of the RNA-dependent RNA polymerase from bovine viral diarrhea virus establishes the role of GTP in de novo initiation.
Proc.Natl.Acad.Sci.Usa, 101, 2004
|
|
8WIE
 
 | |
8WIQ
 
 | |
8WJF
 
 | |
1S9Z
 
 | | SYNTHETIC 17 AMINO ACID LONG PEPTIDE THAT FORMS A NATIVE-LIKE COILED-COIL AT AMBIENT TEMPERATURE AND AGGREGATES INTO AMYLOID-LIKE FIBRILS AT HIGHER TEMPERATURES. | | 分子名称: | SODIUM ION, SYNTHETIC COILED-COIL PEPTIDE, ZINC ION | | 著者 | Kammerer, R.A, Kostrewa, D, Zurdo, J, Detken, A, Garcia-Echeverria, C, Green, J.D, Muller, S.A, Meier, B.H, Winkler, F.K, Dobson, C.M, Steinmetz, M.O. | | 登録日 | 2004-02-06 | | 公開日 | 2004-04-06 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (2.01 Å) | | 主引用文献 | Exploring amyloid formation by a de novo design
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
2L69
 
 | | Solution NMR Structure of de novo designed protein, P-loop NTPase fold, Northeast Structural Genomics Consortium Target OR28 | | 分子名称: | Rossmann 2x3 fold protein | | 著者 | Liu, G, Koga, N, Koga, R, Xiao, R, Mao, A, Mao, B, Patel, D, Ciccosanti, C, Hamilton, K, Acton, T.B, Baker, D, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2010-11-17 | | 公開日 | 2011-01-12 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution NMR structure of de novo designed rossmann 2x3 fold protein, Northeast Structural Genomics Consortium Target OR28
To be Published
|
|
5CIY
 
 | | Structural basis of the recognition of H3K36me3 by DNMT3B PWWP domain | | 分子名称: | DNA (5'-D(P*CP*CP*AP*TP*GP*CP*GP*CP*TP*GP*AP*C)-3'), DNA (5'-D(P*GP*TP*CP*AP*GP*(3DR)P*GP*CP*AP*TP*GP*G)-3'), Modification methylase HhaI, ... | | 著者 | Rondelet, G, DAL MASO, T, Willems, L, Wouters, J. | | 登録日 | 2015-07-13 | | 公開日 | 2016-03-30 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.594 Å) | | 主引用文献 | Structural basis for recognition of histone H3K36me3 nucleosome by human de novo DNA methyltransferases 3A and 3B.
J.Struct.Biol., 194, 2016
|
|
5CIU
 
 | | Structural basis of the recognition of H3K36me3 by DNMT3B PWWP domain | | 分子名称: | DNA (cytosine-5)-methyltransferase 3B, GLYCEROL, Histone H3.2 | | 著者 | Rondelet, G, DAL MASO, T, Willems, L, Wouters, J. | | 登録日 | 2015-07-13 | | 公開日 | 2016-03-30 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.24 Å) | | 主引用文献 | Structural basis for recognition of histone H3K36me3 nucleosome by human de novo DNA methyltransferases 3A and 3B.
J.Struct.Biol., 194, 2016
|
|
5K1S
 
 | | crystal structure of AibC | | 分子名称: | Oxidoreductase, zinc-binding dehydrogenase family, ZINC ION | | 著者 | Bock, T, Mueller, R, Blankenfeldt, W. | | 登録日 | 2016-05-18 | | 公開日 | 2016-08-10 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.55 Å) | | 主引用文献 | Crystal structure of AibC, a reductase involved in alternative de novo isovaleryl coenzyme A biosynthesis in Myxococcus xanthus.
Acta Crystallogr.,Sect.F, 72, 2016
|
|
2LR0
 
 | | Solution NMR structure of de novo designed protein, p-loop ntpase fold, northeast structural genomics consortium target or136 | | 分子名称: | P-loop ntpase fold | | 著者 | Liu, G, Koga, N, Koga, R, Xiao, R, Lee, H, Janjua, H, Kohan, E, Acton, T.B, Everett, J.K, Baker, D, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2012-03-19 | | 公開日 | 2012-07-04 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution NMR structure of de novo designed protein, p-loop ntpase fold, northeast structural genomics consortium target or136
To be Published
|
|
