5VIG
 
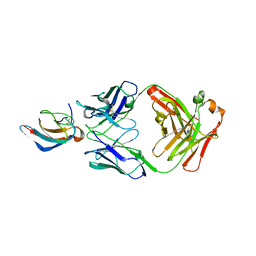 | | Crystal structure of anti-Zika antibody Z006 bound to Zika virus envelope protein DIII | | 分子名称: | CITRATE ANION, Fab heavy chain, Fab light chain, ... | | 著者 | Keeffe, J.R, West Jr, A.P, Gristick, H.B, Bjorkman, P.J. | | 登録日 | 2017-04-16 | | 公開日 | 2017-05-03 | | 最終更新日 | 2017-05-17 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Recurrent Potent Human Neutralizing Antibodies to Zika Virus in Brazil and Mexico.
Cell, 169, 2017
|
|
4P46
 
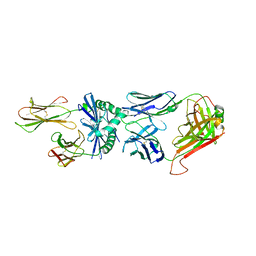 | | J809.B5 Y31A TCR bound to IAb3K | | 分子名称: | 3K Peptide,H-2 class II histocompatibility antigen, A beta chain, H-2 class II histocompatibility antigen, ... | | 著者 | Stadinski, B.D, Huseby, E.S, Trenh, P, Stern, L.J. | | 登録日 | 2014-03-11 | | 公開日 | 2014-05-28 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.851 Å) | | 主引用文献 | Effect of CDR3 Sequences and Distal V Gene Residues in Regulating TCR-MHC Contacts and Ligand Specificity.
J Immunol., 192, 2014
|
|
7V0X
 
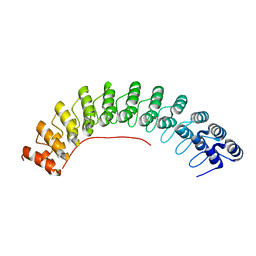 | | Local refinement of ankyrin-1 (C-terminal half), class 1 of erythrocyte ankyrin-1 complex | | 分子名称: | Ankyrin-1 | | 著者 | Vallese, F, Kim, K, Yen, L.Y, Johnston, J.D, Noble, A.J, Cali, T, Clarke, O.B. | | 登録日 | 2022-05-11 | | 公開日 | 2022-07-20 | | 最終更新日 | 2024-02-14 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Architecture of the human erythrocyte ankyrin-1 complex.
Nat.Struct.Mol.Biol., 29, 2022
|
|
1NEK
 
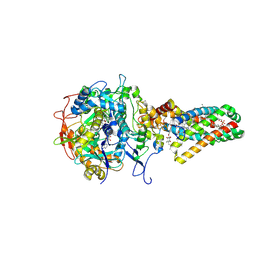 | | Complex II (Succinate Dehydrogenase) From E. Coli with ubiquinone bound | | 分子名称: | CALCIUM ION, CARDIOLIPIN, FE2/S2 (INORGANIC) CLUSTER, ... | | 著者 | Yankovskaya, V, Horsefield, R, Tornroth, S, Luna-Chavez, C, Miyoshi, H, Leger, C, Byrne, B, Cecchini, G, Iwata, S. | | 登録日 | 2002-12-11 | | 公開日 | 2003-02-25 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Architecture of succinate dehydrogenase and
reactive oxygen species generation.
Science, 299, 2003
|
|
1BC2
 
 | |
1XSO
 
 | | THREE-DIMENSIONAL STRUCTURE OF XENOPUS LAEVIS CU,ZN SUPEROXIDE DISMUTASE B DETERMINED BY X-RAY CRYSTALLOGRAPHY AT 1.5 ANGSTROMS RESOLUTION | | 分子名称: | COPPER (II) ION, COPPER,ZINC SUPEROXIDE DISMUTASE, ZINC ION | | 著者 | Djinovic Carugo, K, Coda, A, Battistoni, A, Carri, M.T, Polticelli, F, Desideri, A, Rotilio, G, Wilson, K.S, Bolognesi, M. | | 登録日 | 1995-03-14 | | 公開日 | 1995-07-10 | | 最終更新日 | 2019-08-14 | | 実験手法 | X-RAY DIFFRACTION (1.49 Å) | | 主引用文献 | Three-dimensional structure of Xenopus laevis Cu,Zn superoxide dismutase b determined by X-ray crystallography at 1.5 A resolution.
Acta Crystallogr.,Sect.D, 52, 1996
|
|
1NEN
 
 | | Complex II (Succinate Dehydrogenase) From E. Coli with Dinitrophenol-17 inhibitor co-crystallized at the ubiquinone binding site | | 分子名称: | 2-[1-METHYLHEXYL]-4,6-DINITROPHENOL, CALCIUM ION, CARDIOLIPIN, ... | | 著者 | Yankovskaya, V, Horsefield, R, Tornroth, S, Luna-Chavez, C, Miyoshi, H, Leger, C, Byrne, B, Cecchini, G, Iwata, S. | | 登録日 | 2002-12-11 | | 公開日 | 2003-02-25 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Architecture of succinate dehydrogenase and
reactive oxygen species generation
Science, 299, 2003
|
|
1UY2
 
 | |
5FCZ
 
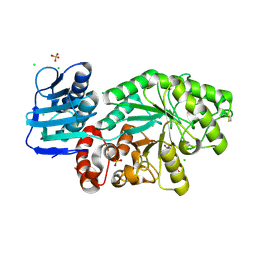 | |
6O2M
 
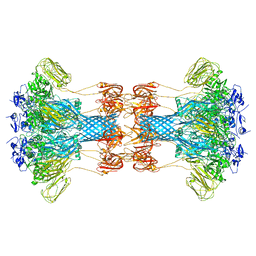 | |
4F27
 
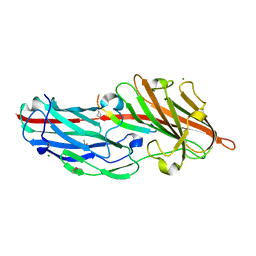 | | Crystal structures reveal the multi-ligand binding mechanism of the Staphylococcus aureus ClfB | | 分子名称: | Clumping factor B, MAGNESIUM ION, peptide from Fibrinogen alpha chain | | 著者 | Yang, M.J, Xiang, H, Wang, J.W, Liu, B, Chen, Y.G, Liu, L, Deng, X.M, Feng, Y. | | 登録日 | 2012-05-07 | | 公開日 | 2012-08-08 | | 最終更新日 | 2015-02-04 | | 実験手法 | X-RAY DIFFRACTION (1.917 Å) | | 主引用文献 | Crystal Structures Reveal the Multi-Ligand Binding Mechanism of Staphylococcus aureus ClfB
Plos Pathog., 8, 2012
|
|
1C11
 
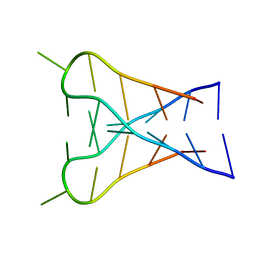 | |
5SQS
 
 | |
5SQR
 
 | |
5SQU
 
 | |
5SR6
 
 | |
5SR5
 
 | |
5SR8
 
 | |
5SRA
 
 | |
5SR9
 
 | |
5SON
 
 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with ZINC000920153280 - (R) isomer | | 分子名称: | 3-{[(2R)-2-phenylpropyl]sulfanyl}-7H-[1,2,4]triazolo[4,3-b][1,2,4]triazole, Non-structural protein 3 | | 著者 | Correy, G.J, Fraser, J.S. | | 登録日 | 2022-06-09 | | 公開日 | 2022-07-13 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.05 Å) | | 主引用文献 | Iterative computational design and crystallographic screening identifies potent inhibitors targeting the Nsp3 macrodomain of SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
5SP3
 
 | |
1DI0
 
 | | CRYSTAL STRUCTURE OF LUMAZINE SYNTHASE FROM BRUCELLA ABORTUS | | 分子名称: | LUMAZINE SYNTHASE, PHOSPHATE ION | | 著者 | Braden, B.C, Velikovsky, C.A, Cauerhff, A.A, Polikarpov, I, Goldbaum, F.A. | | 登録日 | 1999-11-28 | | 公開日 | 2000-04-24 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Divergence in macromolecular assembly: X-ray crystallographic structure analysis of lumazine synthase from Brucella abortus.
J.Mol.Biol., 297, 2000
|
|
5SP1
 
 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with ZINC001472868186 | | 分子名称: | 3-{[methyl(pyrido[2,3-b]pyrazin-6-yl)amino]methyl}[1,2,4]triazolo[4,3-a]pyrazin-8(7H)-one, Non-structural protein 3 | | 著者 | Correy, G.J, Fraser, J.S. | | 登録日 | 2022-06-09 | | 公開日 | 2022-07-13 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.03 Å) | | 主引用文献 | Iterative computational design and crystallographic screening identifies potent inhibitors targeting the Nsp3 macrodomain of SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
5SOJ
 
 | |
