3RBF
 
 | | Crystal structure of Human aromatic L-amino acid decarboxylase (AADC) in the apo form | | 分子名称: | Aromatic-L-amino-acid decarboxylase, CHLORIDE ION, PYRIDOXAL-5'-PHOSPHATE | | 著者 | Giardina, G, Montioli, R, Gianni, S, Cellini, B, Paiardini, A, Borri Voltattorni, C, Cutruzzola, F. | | 登録日 | 2011-03-29 | | 公開日 | 2011-10-19 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Open conformation of human DOPA decarboxylase reveals the mechanism of PLP addition to Group II decarboxylases.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
4E1O
 
 | | Human histidine decarboxylase complex with Histidine methyl ester (HME) | | 分子名称: | HISTIDINE-METHYL-ESTER, Histidine decarboxylase, PYRIDOXAL-5'-PHOSPHATE | | 著者 | Komori, H, Nitta, Y, Ueno, H, Higuchi, Y. | | 登録日 | 2012-03-06 | | 公開日 | 2012-07-18 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structural study reveals that Ser-354 determines substrate specificity on human histidine decarboxylase
J.Biol.Chem., 287, 2012
|
|
3VP6
 
 | | Structural characterization of Glutamic Acid Decarboxylase; insights into the mechanism of autoinactivation | | 分子名称: | 4-oxo-4H-pyran-2,6-dicarboxylic acid, GLYCEROL, Glutamate decarboxylase 1 | | 著者 | Langendorf, C.G, Tuck, K.L, Key, T.L.G, Rosado, C.J, Wong, A.S.M, Fenalti, G, Buckle, A.M, Law, R.H.P, Whisstock, J.C. | | 登録日 | 2012-02-27 | | 公開日 | 2013-01-16 | | 最終更新日 | 2013-08-14 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structural characterization of the mechanism through which human glutamic acid decarboxylase auto-activates
Biosci.Rep., 33, 2013
|
|
4Q6R
 
 | |
4RIT
 
 | | The yellow crystal structure of pyridoxal-dependent decarboxylase from sphaerobacter thermophilus dsm 20745 | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, GLYCEROL, ... | | 著者 | Wu, R, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2014-10-07 | | 公開日 | 2014-10-22 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | The yellow crystal structure of pyridoxal-dependent decarboxylase from sphaerobacter thermophilus dsm 20745
To be Published
|
|
4OBU
 
 | |
4RLG
 
 | |
4OBV
 
 | | Ruminococcus gnavus tryptophan decarboxylase RUMGNA_01526 (alpha-FMT) | | 分子名称: | Pyridoxal-dependent decarboxylase domain protein, alpha-(fluoromethyl)-D-tryptophan, {5-hydroxy-4-[(1E)-4-(1H-indol-3-yl)-3-oxobut-1-en-1-yl]-6-methylpyridin-3-yl}methyl dihydrogen phosphate | | 著者 | Fraser, J.S, Van Benschoten, A.H. | | 登録日 | 2014-01-07 | | 公開日 | 2014-10-29 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.84 Å) | | 主引用文献 | Discovery and Characterization of Gut Microbiota Decarboxylases that Can Produce the Neurotransmitter Tryptamine.
Cell Host Microbe, 16, 2014
|
|
4W8I
 
 | | Crystal structure of LpSPL/Lpp2128, Legionella pneumophila sphingosine-1 phosphate lyase | | 分子名称: | Probable sphingosine-1-phosphate lyase | | 著者 | Stogios, P.J, Daniels, C, Skarina, T, Cuff, M, Di Leo, R, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2014-08-24 | | 公開日 | 2014-11-05 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.85 Å) | | 主引用文献 | Legionella pneumophila S1P-lyase targets host sphingolipid metabolism and restrains autophagy.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
4RM1
 
 | |
4RIZ
 
 | |
4RJ0
 
 | |
5EUE
 
 | | S1P Lyase Bacterial Surrogate bound to N-(2-((4-methoxy-2,5-dimethylbenzyl)amino)-1-phenylethyl)-5-methylisoxazole-3-carboxamide | | 分子名称: | PHOSPHATE ION, Putative sphingosine-1-phosphate lyase, ~{N}-[(1~{S})-2-[(4-methoxy-2,5-dimethyl-phenyl)methylamino]-1-phenyl-ethyl]-5-methyl-1,2-oxazole-3-carboxamide | | 著者 | Argiriadi, M.A, Banach, D, Radziejewska, E, Marchie, S, DiMauro, J, Dinges, J, Dominguez, E, Hutchins, C, Judge, R.A, Queeney, K, Wallace, G, Harris, C.M. | | 登録日 | 2015-11-18 | | 公開日 | 2016-03-16 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.83 Å) | | 主引用文献 | Creation of a S1P Lyase bacterial surrogate for structure-based drug design.
Bioorg.Med.Chem.Lett., 26, 2016
|
|
5EUD
 
 | | S1P Lyase Bacterial Surrogate bound to N-(1-(4-(3-hydroxyprop-1-yn-1-yl)phenyl)-2-((4-methoxy-2,5-dimethylbenzyl)amino)ethyl)-5-methylisoxazole-3-carboxamide | | 分子名称: | PHOSPHATE ION, Putative sphingosine-1-phosphate lyase, ~{N}-[(1~{S})-2-[(4-methoxy-2,5-dimethyl-phenyl)methylamino]-1-[4-(3-oxidanylprop-1-ynyl)phenyl]ethyl]-5-methyl-1,2-oxazole-3-carboxamide | | 著者 | Argiriadi, M.A, Banach, D, Radziejewska, E, Marchie, S, DiMauro, J, Dinges, J, Dominguez, E, Hutchins, C, Judge, R.A, Queeney, K, Wallace, G, Harris, C.M. | | 登録日 | 2015-11-18 | | 公開日 | 2016-03-16 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.24 Å) | | 主引用文献 | Creation of a S1P Lyase bacterial surrogate for structure-based drug design.
Bioorg.Med.Chem.Lett., 26, 2016
|
|
5HSJ
 
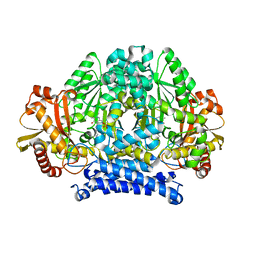 | | Structure of tyrosine decarboxylase complex with PLP at 1.9 Angstroms resolution | | 分子名称: | PYRIDOXAL-5'-PHOSPHATE, Putative decarboxylase | | 著者 | Ni, Y, Zhou, J, Zhu, H, Zhang, K. | | 登録日 | 2016-01-25 | | 公開日 | 2016-09-14 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystal structure of tyrosine decarboxylase and identification of key residues involved in conformational swing and substrate binding
Sci Rep, 6, 2016
|
|
5HSI
 
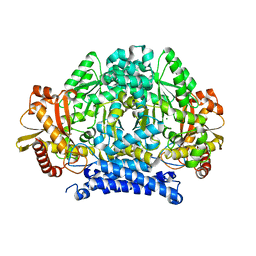 | | Crystal structure of tyrosine decarboxylase at 1.73 Angstroms resolution | | 分子名称: | MAGNESIUM ION, Putative decarboxylase | | 著者 | Ni, Y, Zhou, J, Zhu, H, Zhang, K. | | 登録日 | 2016-01-25 | | 公開日 | 2016-09-21 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.732 Å) | | 主引用文献 | Crystal structure of tyrosine decarboxylase and identification of key residues involved in conformational swing and substrate binding
Sci Rep, 6, 2016
|
|
5K1R
 
 | |
5GP4
 
 | |
6ENZ
 
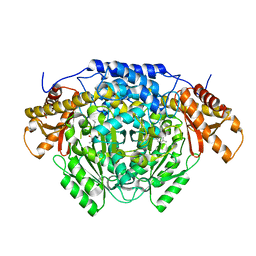 | | Crystal structure of mouse GADL1 | | 分子名称: | Acidic amino acid decarboxylase GADL1, PYRIDOXAL-5'-PHOSPHATE | | 著者 | Raasakka, A, Mahootchi, E, Winge, I, Luan, W, Kursula, P, Haavik, J. | | 登録日 | 2017-10-07 | | 公開日 | 2018-01-03 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Structure of the mouse acidic amino acid decarboxylase GADL1.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
5O5C
 
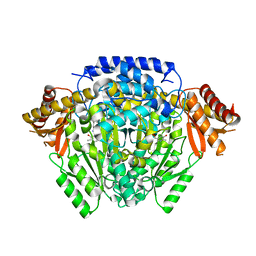 | | The crystal structure of DfoJ, the desferrioxamine biosynthetic pathway lysine decarboxylase from the fire blight disease pathogen Erwinia amylovora | | 分子名称: | PYRIDOXAL-5'-PHOSPHATE, Putative decarboxylase involved in desferrioxamine biosynthesis | | 著者 | Salomone-Stagni, M, Bartho, J.D, Polsinelli, I, Bellini, D, Walsh, M.A, Demitri, N, Benini, S. | | 登録日 | 2017-06-01 | | 公開日 | 2018-02-28 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | A complete structural characterization of the desferrioxamine E biosynthetic pathway from the fire blight pathogen Erwinia amylovora.
J. Struct. Biol., 202, 2018
|
|
6EEM
 
 | | Crystal structure of Papaver somniferum tyrosine decarboxylase in complex with L-tyrosine | | 分子名称: | N-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methyl)-L-tyrosine, SULFATE ION, TYROSINE, ... | | 著者 | Torrens-Spence, M.P, Chiang, Y, Smith, T, Vicent, M.A, Wang, Y, Weng, J.K. | | 登録日 | 2018-08-14 | | 公開日 | 2018-09-19 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.61000657 Å) | | 主引用文献 | Structural basis for divergent and convergent evolution of catalytic machineries in plant aromatic amino acid decarboxylase proteins.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6EEQ
 
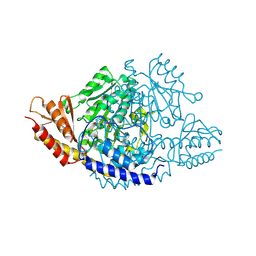 | | Crystal structure of Rhodiola rosea 4-hydroxyphenylacetaldehyde synthase | | 分子名称: | 4-hydroxyphenylacetaldehyde synthase | | 著者 | Torrens-Spence, M.P, Chiang, Y, Smith, T, Vicent, M.A, Wang, Y, Weng, J.K. | | 登録日 | 2018-08-15 | | 公開日 | 2018-09-19 | | 最終更新日 | 2020-06-03 | | 実験手法 | X-RAY DIFFRACTION (2.600086 Å) | | 主引用文献 | Structural basis for divergent and convergent evolution of catalytic machineries in plant aromatic amino acid decarboxylase proteins.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6EEI
 
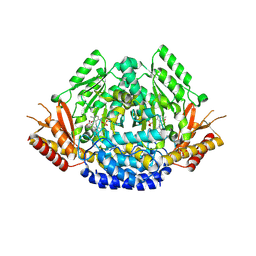 | | Crystal structure of Arabidopsis thaliana phenylacetaldehyde synthase in complex with L-phenylalanine | | 分子名称: | PHENYLALANINE, SULFATE ION, Tyrosine decarboxylase 1 | | 著者 | Torrens-Spence, M.P, Chiang, Y, Smith, T, Vicent, M.A, Wang, Y, Weng, J.K. | | 登録日 | 2018-08-14 | | 公開日 | 2018-09-19 | | 最終更新日 | 2020-06-03 | | 実験手法 | X-RAY DIFFRACTION (1.99001348 Å) | | 主引用文献 | Structural basis for divergent and convergent evolution of catalytic machineries in plant aromatic amino acid decarboxylase proteins.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6EEW
 
 | | Crystal structure of Catharanthus roseus tryptophan decarboxylase in complex with L-tryptophan | | 分子名称: | Aromatic-L-amino-acid decarboxylase, CALCIUM ION, TRYPTOPHAN | | 著者 | Torrens-Spence, M.P, Chiang, Y, Smith, T, Vicent, M.A, Wang, Y, Weng, J.K. | | 登録日 | 2018-08-15 | | 公開日 | 2018-09-19 | | 最終更新日 | 2020-06-03 | | 実験手法 | X-RAY DIFFRACTION (2.05002069 Å) | | 主引用文献 | Structural basis for divergent and convergent evolution of catalytic machineries in plant aromatic amino acid decarboxylase proteins.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6EBN
 
 | | Crystal structure of Psilocybe cubensis noncanonical aromatic amino acid decarboxylase | | 分子名称: | FORMIC ACID, GLYCEROL, SODIUM ION, ... | | 著者 | Torrens-Spence, M.P, Chun-Ting, L, Pluskal, T, Chung, Y.K, Weng, J.K. | | 登録日 | 2018-08-06 | | 公開日 | 2018-12-12 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.9663111 Å) | | 主引用文献 | Monoamine Biosynthesis via a Noncanonical Calcium-Activatable Aromatic Amino Acid Decarboxylase in Psilocybin Mushroom.
ACS Chem. Biol., 13, 2018
|
|
