1AT6
 
 | | HEN EGG WHITE LYSOZYME WITH A ISOASPARTATE RESIDUE | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, LYSOZYME | | 著者 | Noguchi, S, Miyawaki, K, Satow, Y. | | 登録日 | 1997-08-19 | | 公開日 | 1998-02-25 | | 最終更新日 | 2024-11-20 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Succinimide and isoaspartate residues in the crystal structures of hen egg-white lysozyme complexed with tri-N-acetylchitotriose.
J.Mol.Biol., 278, 1998
|
|
6HT0
 
 | | Crystal structure of MLLT1 (ENL) YEATS domain in complexed with compound 94 | | 分子名称: | 1,2-ETHANEDIOL, 1-cyclopropyl-~{N}-[2-[[(2~{S})-2-methylpyrrolidin-1-yl]methyl]-3~{H}-benzimidazol-5-yl]indazole-5-carboxamide, Protein ENL, ... | | 著者 | Heidenreich, D, Chaikuad, A, Moustakim, M, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Fedorov, O, Brennan, P.E, Knapp, S, Structural Genomics Consortium (SGC) | | 登録日 | 2018-10-02 | | 公開日 | 2018-10-17 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Discovery of an MLLT1/3 YEATS Domain Chemical Probe.
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
8S3X
 
 | | LIM Domain Kinase 2 (LIMK2) bound to compound 52 | | 分子名称: | 4-(5-cyclopropyl-7~{H}-pyrrolo[2,3-d]pyrimidin-4-yl)-~{N}-[3-(3-methoxyphenyl)phenyl]-3,6-dihydro-2~{H}-pyridine-1-carboxamide, LIM domain kinase 2 | | 著者 | Mathea, S, Chatterjee, D, Preuss, F, Ple, K, Knapp, S. | | 登録日 | 2024-02-20 | | 公開日 | 2024-03-06 | | 最終更新日 | 2024-09-18 | | 実験手法 | X-RAY DIFFRACTION (2.59 Å) | | 主引用文献 | Tetrahydropyridine LIMK inhibitors: Structure activity studies and biological characterization.
Eur.J.Med.Chem., 271, 2024
|
|
5IIS
 
 | | Design, synthesis and structure activity relationship of potent pan-PIM kinase inhibitors derived from the pyridyl-amide scaffold | | 分子名称: | 3-amino-N-(2'-amino-6'-methyl[4,4'-bipyridin]-3-yl)-6-(2-fluorophenyl)pyridine-2-carboxamide, DI(HYDROXYETHYL)ETHER, Serine/threonine-protein kinase pim-1 | | 著者 | Bellamacina, C, Bussiere, D, Burger, M. | | 登録日 | 2016-03-01 | | 公開日 | 2016-04-06 | | 最終更新日 | 2024-11-20 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Design, synthesis and structure activity relationship of potent pan-PIM kinase inhibitors derived from the pyridyl carboxamide scaffold.
Bioorg.Med.Chem.Lett., 26, 2016
|
|
8S1K
 
 | | Crystal Structure of human FABP4 in complex with 2-[1-(methoxymethyl)cyclopentyl]-6-pentyl-4-phenyl-3-(1H-tetrazol-5-yl)-5,6,7,8-tetrahydroquinoline | | 分子名称: | (6~{S})-2-[1-(methoxymethyl)cyclopentyl]-6-pentyl-4-phenyl-3-(1~{H}-1,2,3,4-tetrazol-5-yl)-5,6,7,8-tetrahydroquinoline, DIMETHYL SULFOXIDE, Fatty acid-binding protein, ... | | 著者 | Ehler, A, Benz, J, Obst-Sander, U, Rudolph, M.G. | | 登録日 | 2024-02-15 | | 公開日 | 2024-03-06 | | 最終更新日 | 2025-09-17 | | 実験手法 | X-RAY DIFFRACTION (1.22 Å) | | 主引用文献 | A high-resolution data set of fatty acid-binding protein structures. I. Dynamics of FABP4 and ligand binding.
Acta Crystallogr D Struct Biol, 81, 2025
|
|
5WY8
 
 | | Crystal structure of PTP delta Ig1-Ig3 in complex with IL1RAPL1 Ig1-Ig3 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Interleukin-1 receptor accessory protein-like 1, ... | | 著者 | Kim, H.M, Won, S.Y, Kim, D. | | 登録日 | 2017-01-11 | | 公開日 | 2017-11-22 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (3.07 Å) | | 主引用文献 | LAR-RPTP Clustering Is Modulated by Competitive Binding between Synaptic Adhesion Partners and Heparan Sulfate
Front Mol Neurosci, 10, 2017
|
|
5FQA
 
 | | Crystal Structure of Bacillus cereus Metallo-Beta-Lactamase II | | 分子名称: | 1,2-ETHANEDIOL, BETA-LACTAMASE 2, FE (III) ION, ... | | 著者 | Cahill, S.T, Brem, J, McDonough, M.A, Schofield, C.J. | | 登録日 | 2015-12-08 | | 公開日 | 2016-08-10 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (1.1 Å) | | 主引用文献 | Use of ferrous iron by metallo-beta-lactamases.
J. Inorg. Biochem., 163, 2016
|
|
8XOA
 
 | | VMAT2 complex with MPP+ | | 分子名称: | 1-methyl-4-phenylpyridin-1-ium, Synaptic vesicular amine transporter,Synaptic vesicular amine transporter,transporterA | | 著者 | Jiang, D.H, Wu, D. | | 登録日 | 2023-12-31 | | 公開日 | 2024-05-08 | | 最終更新日 | 2025-06-18 | | 実験手法 | ELECTRON MICROSCOPY (3.03 Å) | | 主引用文献 | Structural snapshots of human VMAT2 reveal insights into substrate recognition and proton coupling mechanism.
Cell Res., 34, 2024
|
|
5IS5
 
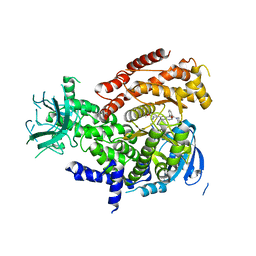 | |
1B1Y
 
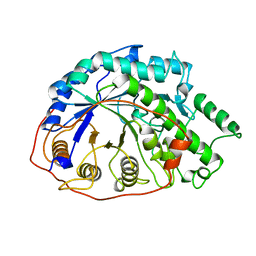 | | SEVENFOLD MUTANT OF BARLEY BETA-AMYLASE | | 分子名称: | PROTEIN (BETA-AMYLASE), alpha-D-glucopyranose-(1-4)-beta-D-glucopyranose, beta-D-glucopyranose | | 著者 | Mikami, B, Yoon, H.J, Yoshigi, N. | | 登録日 | 1998-11-25 | | 公開日 | 1998-12-02 | | 最終更新日 | 2023-08-09 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | The crystal structure of the sevenfold mutant of barley beta-amylase with increased thermostability at 2.5 A resolution.
J.Mol.Biol., 285, 1999
|
|
5IQ6
 
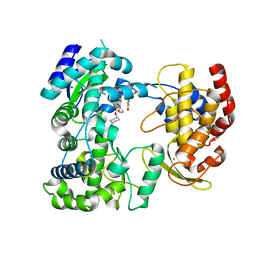 | | Crystal structure of Dengue virus serotype 3 RNA dependent RNA polymerase bound to HeE1-2Tyr, a new pyridobenzothizole inhibitor | | 分子名称: | N-[8-(cyclohexyloxy)-1-oxo-2-phenyl-1H-pyrido[2,1-b][1,3]benzothiazole-4-carbonyl]-L-tyrosine, RNA dependent RNA polymerase, ZINC ION | | 著者 | Tarantino, D, Mastrangelo, E, Milani, M. | | 登録日 | 2016-03-10 | | 公開日 | 2016-10-05 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Targeting flavivirus RNA dependent RNA polymerase through a pyridobenzothiazole inhibitor.
Antiviral Res., 134, 2016
|
|
4MUW
 
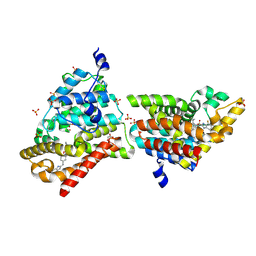 | | Crystal Structure of PDE10A with Novel Keto-Benzimidazole Inhibitor | | 分子名称: | 2-{4-[(6,7-difluoro-1H-benzimidazol-2-yl)amino]phenoxy}-N-methyl-3,4'-bipyridin-2'-amine, GLYCEROL, SULFATE ION, ... | | 著者 | Chmait, S, Jordan, S. | | 登録日 | 2013-09-23 | | 公開日 | 2013-10-23 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (2.639 Å) | | 主引用文献 | Design, Optimization, and Biological Evaluation of Novel Keto-Benzimidazoles as Potent and Selective Inhibitors of Phosphodiesterase 10A (PDE10A).
J.Med.Chem., 56, 2013
|
|
6HUZ
 
 | | HmdII from Desulfurobacterium thermolithotrophum reconstituted with Fe-guanylylpyridinol (FeGP) cofactor and co-crystallized with methenyl-tetrahydrofolate form B | | 分子名称: | 1,2-ETHANEDIOL, 5,10-Methenyltetrahydrofolate, Coenzyme F420-dependent N(5),N(10)-methenyltetrahydromethanopterin reductase-related protein, ... | | 著者 | Watanabe, T, Wagner, T, Huang, G, Kahnt, J, Ataka, K, Ermler, U, Shima, S. | | 登録日 | 2018-10-09 | | 公開日 | 2019-01-09 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | The Bacterial [Fe]-Hydrogenase Paralog HmdII Uses Tetrahydrofolate Derivatives as Substrates.
Angew. Chem. Int. Ed. Engl., 58, 2019
|
|
8S0J
 
 | | A fragment-based inhibitor of SHP2 | | 分子名称: | 3-[3-[2,3-bis(chloranyl)phenyl]-1H-pyrrolo[3,2-b]pyridin-6-yl]propan-1-amine, Tyrosine-protein phosphatase non-receptor type 11 | | 著者 | Cleasby, A, Price, A. | | 登録日 | 2024-02-14 | | 公開日 | 2024-03-20 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (1.89 Å) | | 主引用文献 | Fragment-Based Discovery of Allosteric Inhibitors of SH2 Domain-Containing Protein Tyrosine Phosphatase-2 (SHP2).
J.Med.Chem., 67, 2024
|
|
3MAM
 
 | | A molecular switch changes the low to the high affinity state in the substrate binding protein AfProX | | 分子名称: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Osmoprotection protein (ProX), ... | | 著者 | Tschapek, B, Pittelkow, M, Bremer, E, Schmitt, L, Smits, S.H. | | 登録日 | 2010-03-24 | | 公開日 | 2011-04-06 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Arg149 Is Involved in Switching the Low Affinity, Open State of the Binding Protein AfProX into Its High Affinity, Closed State.
J.Mol.Biol., 411, 2011
|
|
6HX7
 
 | | Crystal structure of human R180T variant of ORNITHINE AMINOTRANSFERASE at 1.8 Angstrom | | 分子名称: | Ornithine aminotransferase, mitochondrial, PYRIDOXAL-5'-PHOSPHATE | | 著者 | Giardina, G, Montioli, R, Cellini, B, Cutruzzola, F, Borri Voltattorni, C. | | 登録日 | 2018-10-16 | | 公開日 | 2019-06-05 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | R180T variant of delta-ornithine aminotransferase associated with gyrate atrophy: biochemical, computational, X-ray and NMR studies provide insight into its catalytic features.
Febs J., 286, 2019
|
|
8RX9
 
 | | LTA4 hydrolase in complex with compound3 | | 分子名称: | 1-[[5-[5-(1~{H}-pyrazol-5-yl)pyridin-2-yl]oxypyridin-2-yl]methyl]piperidin-4-ol, ACETATE ION, IMIDAZOLE, ... | | 著者 | Srinivas, H. | | 登録日 | 2024-02-06 | | 公開日 | 2024-03-20 | | 最終更新日 | 2024-04-10 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Structure-Guided Elaboration of a Fragment-Like Hit into an Orally Efficacious Leukotriene A4 Hydrolase Inhibitor.
J.Med.Chem., 67, 2024
|
|
5IDQ
 
 | |
9HNL
 
 | | Structure of the (6-4) photolyase of Caulobacter crescentus in its oxidized state at room temperture-synchrotron | | 分子名称: | 1-deoxy-1-(6,7-dimethyl-2,4-dioxo-3,4-dihydropteridin-8(2H)-yl)-D-ribitol, Cryptochrome/photolyase family protein, FLAVIN-ADENINE DINUCLEOTIDE, ... | | 著者 | Po Hsun, W, Maestre-Reyna, M, Essen, L.-O. | | 登録日 | 2024-12-11 | | 公開日 | 2025-05-07 | | 最終更新日 | 2025-05-21 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Redox-State-Dependent Structural Changes within a Prokaryotic 6-4 Photolyase.
J.Am.Chem.Soc., 147, 2025
|
|
7RHC
 
 | | A new fluorescent protein darkmRuby at pH 9.0 | | 分子名称: | 1,2-ETHANEDIOL, darkmRuby | | 著者 | Huang, M, Ng, H.L, Zhang, S, Deng, M, Chu, J. | | 登録日 | 2021-07-16 | | 公開日 | 2022-07-27 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | A Long-range Interaction Affects Brightness and pH Stability of a Dark Fluorescent Protein
To Be Published
|
|
6HSW
 
 | | A CE15 glucuronoyl esterase from Teredinibacter turnerae T7901 | | 分子名称: | 1,2-ETHANEDIOL, BROMIDE ION, Carbohydrate esterase family 15 domain protein, ... | | 著者 | Mazurkewich, S, Lo Leggio, L, Navarro Poulsen, J.C, Larsbrink, J. | | 登録日 | 2018-10-02 | | 公開日 | 2019-03-20 | | 最終更新日 | 2024-11-13 | | 実験手法 | X-RAY DIFFRACTION (2.14734387 Å) | | 主引用文献 | Structure-function analyses reveal that a glucuronoyl esterase fromTeredinibacter turneraeinteracts with carbohydrates and aromatic compounds.
J.Biol.Chem., 294, 2019
|
|
3MPD
 
 | |
7Y0L
 
 | | SulE-S209A | | 分子名称: | Alpha/beta fold hydrolase, GLYCEROL, METHYL 2-[({[(4-METHOXY-6-METHYL-1,3,5-TRIAZIN-2-YL)AMINO]CARBONYL}AMINO)SULFONYL]BENZOATE | | 著者 | Liu, B, Ran, T, He, J, Wang, W. | | 登録日 | 2022-06-05 | | 公開日 | 2023-08-02 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (1.29 Å) | | 主引用文献 | Crystal structures of herbicide-detoxifying esterase reveal a lid loop affecting substrate binding and activity.
Nat Commun, 14, 2023
|
|
8S6H
 
 | |
9E6I
 
 | | Cryo-EM structure of Saccharomyces cerevisiae Pmt4-Ccw5 complex | | 分子名称: | (3R)-3,31-dimethyl-7,11,15,19,23,27-hexamethylidenedotriacont-31-en-1-yl dihydrogen phosphate, Cell wall mannoprotein CIS3, Dolichyl-phosphate-mannose--protein mannosyltransferase 4 | | 著者 | Du, M, Yuan, Z, Li, H. | | 登録日 | 2024-10-30 | | 公開日 | 2025-10-15 | | 最終更新日 | 2025-11-19 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Pmt4 recognizes two separate acceptor sites to O-mannosylate in the S/T-rich regions of substrate proteins.
Nat Commun, 16, 2025
|
|
