5YQT
 
 | | Crystal Structure of the L74F/M78V/I80V/L114F mutant of LEH complexed with cyclopentene oxide | | 分子名称: | (1R,5S)-6-oxabicyclo[3.1.0]hexane, Limonene-1,2-epoxide hydrolase | | 著者 | Kong, X.D, Sun, Z.T, Wu, L, Reetz, M.T, Zhou, J.H, Xu, J.H. | | 登録日 | 2017-11-07 | | 公開日 | 2018-06-27 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structural and Computational Insight into the Catalytic Mechanism of Limonene Epoxide Hydrolase Mutants in Stereoselective Transformations.
J. Am. Chem. Soc., 140, 2018
|
|
3RWL
 
 | | Structure of P450pyr hydroxylase | | 分子名称: | Cytochrome P450 alkane hydroxylase 1 CYP153A7, PROTOPORPHYRIN IX CONTAINING FE | | 著者 | Pompidor, G. | | 登録日 | 2011-05-09 | | 公開日 | 2012-04-18 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Evolving P450pyr hydroxylase for highly enantioselective hydroxylation at non-activated carbon atom.
Chem.Commun.(Camb.), 48, 2012
|
|
5IKX
 
 | |
8PEC
 
 | | OXA-48_Q5-CAZ. Epistasis Arises from Shifting the Rate-Limiting Step during Enzyme Evolution | | 分子名称: | 1-({(2R)-2-[(1R)-1-{[(2Z)-2-(2-amino-1,3-thiazol-4-yl)-2-{[(2-carboxypropan-2-yl)oxy]imino}acetyl]amino}-2-oxoethyl]-4-carboxy-3,6-dihydro-2H-1,3-thiazin-5-yl}methyl)pyridinium, Beta-lactamase, CHLORIDE ION | | 著者 | Leiros, H.-K.S, Frohlich, C. | | 登録日 | 2023-06-13 | | 公開日 | 2024-02-14 | | 最終更新日 | 2024-06-12 | | 実験手法 | X-RAY DIFFRACTION (2.66 Å) | | 主引用文献 | Epistasis arises from shifting the rate-limiting step during enzyme evolution of a beta-lactamase.
Nat Catal, 7, 2024
|
|
8PEB
 
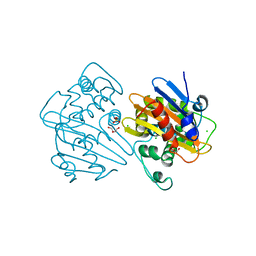 | |
8PEA
 
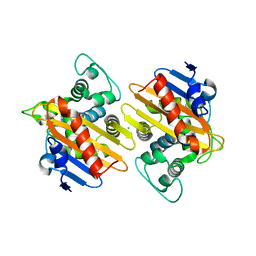 | |
8ZF1
 
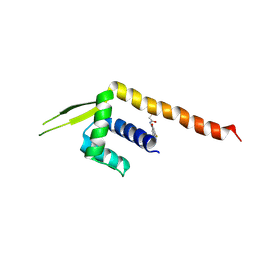 | | Crystal structure of a Chemo Triplet Photoenzyme (CTPe) | | 分子名称: | N-(9-oxidanylidenethioxanthen-2-yl)ethanamide, Transcriptional regulator, PadR-like family | | 著者 | Chen, X, Qian, J.Y, Guo, J, Wu, Y.Z, Zhong, F.R. | | 登録日 | 2024-05-07 | | 公開日 | 2024-07-17 | | 最終更新日 | 2024-07-24 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Chemogenetic Evolution of Diversified Photoenzymes for Enantioselective [2 + 2] Cycloadditions in Whole Cells.
J.Am.Chem.Soc., 146, 2024
|
|
1U3Y
 
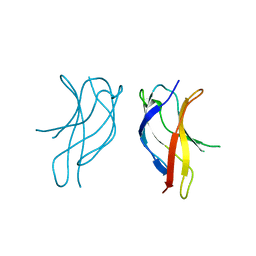 | | Crystal structure of ILAC mutant of dimerisation domain of NF-kB p50 transcription factor | | 分子名称: | Nuclear factor NF-kappa-B p105 subunit | | 著者 | Chirgadze, D.Y, Demydchuk, M, Becker, M, Moran, S, Paoli, M. | | 登録日 | 2004-07-22 | | 公開日 | 2004-08-17 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (1.901 Å) | | 主引用文献 | Snapshot of Protein Structure Evolution Reveals Conservation of Functional Dimerization through Intertwined Folding
Structure, 12, 2004
|
|
1U42
 
 | | Crystal structure of MLAM mutant of dimerisation domain of NF-kB p50 transcription factor | | 分子名称: | Nuclear factor NF-kappa-B p105 subunit | | 著者 | Chirgadze, D.Y, Demydchuk, M, Becker, M, Moran, S, Paoli, M. | | 登録日 | 2004-07-23 | | 公開日 | 2004-08-17 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.699 Å) | | 主引用文献 | Snapshot of Protein Structure Evolution Reveals Conservation of Functional Dimerization through Intertwined Folding
Structure, 12, 2004
|
|
1U3Z
 
 | | Crystal structure of MLAC mutant of dimerisation domain of NF-kB p50 transcription factor | | 分子名称: | Nuclear factor NF-kappa-B p105 subunit | | 著者 | Chirgadze, D.Y, Demydchuk, M, Becker, M, Moran, S, Paoli, M. | | 登録日 | 2004-07-23 | | 公開日 | 2004-08-17 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Snapshot of Protein Structure Evolution Reveals Conservation of Functional Dimerization through Intertwined Folding
Structure, 12, 2004
|
|
1U3J
 
 | | Crystal structure of MLAV mutant of dimerisation domain of NF-kB p50 transcription factor | | 分子名称: | Nuclear factor NF-kappa-B p105 subunit | | 著者 | Chirgadze, D.Y, Demydchuk, M, Becker, M, Moran, S, Paoli, M. | | 登録日 | 2004-07-22 | | 公開日 | 2004-08-17 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (1.901 Å) | | 主引用文献 | Snapshot of Protein Structure Evolution Reveals Conservation of Functional Dimerization through Intertwined Folding
Structure, 12, 2004
|
|
2QBW
 
 | | The crystal structure of PDZ-Fibronectin fusion protein | | 分子名称: | PDZ-Fibronectin fusion protein, Polypeptide | | 著者 | Huang, J, Makabe, K, Koide, A, Koide, S. | | 登録日 | 2007-06-18 | | 公開日 | 2008-04-22 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Design of protein function leaps by directed domain interface evolution.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
2P3S
 
 | | Crystal structure of a thermostable mutant of Bacillus subtilis Adenylate Kinase (G214R/Q199R) | | 分子名称: | Adenylate kinase, BIS(ADENOSINE)-5'-PENTAPHOSPHATE, MAGNESIUM ION, ... | | 著者 | Counago, R, Wilson, C.J, Myers, J, Wu, G, Shamoo, Y. | | 登録日 | 2007-03-09 | | 公開日 | 2008-01-22 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal structure of a thermostable mutant of Bacillus subtilis Adenylate Kinase (G214R/Q199R)
To be Published
|
|
3UZ5
 
 | | Designed protein KE59 R13 3/11H | | 分子名称: | 5,7-dichloro-1H-benzotriazole, Kemp eliminase KE59 R13 3/11H, PHOSPHATE ION, ... | | 著者 | Khersonsky, O, Kiss, G, Roethlisberger, D, Dym, O, Albeck, S, Houk, K.N, Baker, D, Tawfik, D.S, Israel Structural Proteomics Center (ISPC) | | 登録日 | 2011-12-07 | | 公開日 | 2012-06-06 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Bridging the gaps in design methodologies by evolutionary optimization of the stability and proficiency of designed Kemp eliminase KE59.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3UZJ
 
 | | Designed protein KE59 R13 3/11H with benzotriazole | | 分子名称: | 1H-benzotriazole, Kemp eliminase KE59 R13 3/11H, PHOSPHATE ION | | 著者 | Khersonsky, O, Kiss, G, Roethlisberger, D, Dym, O, Albeck, S, Houk, K.N, Baker, D, Tawfik, D.S, Israel Structural Proteomics Center (ISPC) | | 登録日 | 2011-12-07 | | 公開日 | 2012-06-06 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.69 Å) | | 主引用文献 | Bridging the gaps in design methodologies by evolutionary optimization of the stability and proficiency of designed Kemp eliminase KE59.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3UYC
 
 | | Designed protein KE59 R8_2/7A | | 分子名称: | Kemp eliminase KE59 R8_2/7A, PHOSPHATE ION | | 著者 | Khersonsky, O, Kiss, G, Roethlisberger, D, Dym, O, Albeck, S, Houk, K.N, Baker, D, Tawfik, D.S, Israel Structural Proteomics Center (ISPC) | | 登録日 | 2011-12-06 | | 公開日 | 2012-06-06 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Bridging the gaps in design methodologies by evolutionary optimization of the stability and proficiency of designed Kemp eliminase KE59.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3UXD
 
 | | Designed protein KE59 R1 7/10H with dichlorobenzotriazole (DBT) | | 分子名称: | 5,7-dichloro-1H-benzotriazole, Kemp eliminase KE59 R1 7/10H, PHOSPHATE ION | | 著者 | Khersonsky, O, Kiss, G, Roethlisberger, D, Dym, O, Albeck, S, Houk, K.N, Baker, D, Tawfik, D.S, Israel Structural Proteomics Center (ISPC) | | 登録日 | 2011-12-05 | | 公開日 | 2012-06-06 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Bridging the gaps in design methodologies by evolutionary optimization of the stability and proficiency of designed Kemp eliminase KE59.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3UY7
 
 | | Designed protein KE59 R1 7/10H with G130S mutation | | 分子名称: | Kemp eliminase KE59 R1 7/10H, SODIUM ION, SULFATE ION | | 著者 | Khersonsky, O, Kiss, G, Roethlisberger, D, Dym, O, Albeck, S, Houk, K.N, Baker, D, Tawfik, D.S, Israel Structural Proteomics Center (ISPC) | | 登録日 | 2011-12-06 | | 公開日 | 2012-06-27 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Bridging the gaps in design methodologies by evolutionary optimization of the stability and proficiency of designed Kemp eliminase KE59.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3UXA
 
 | | Designed protein KE59 R1 7/10H | | 分子名称: | Kemp eliminase KE59 R1 7/10H, PHOSPHATE ION | | 著者 | Khersonsky, O, Kiss, G, Roethlisberger, D, Dym, O, Albeck, S, Houk, K.N, Baker, D, Tawfik, D.S, Israel Structural Proteomics Center (ISPC) | | 登録日 | 2011-12-05 | | 公開日 | 2012-06-06 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Bridging the gaps in design methodologies by evolutionary optimization of the stability and proficiency of designed Kemp eliminase KE59.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3UY8
 
 | | Designed protein KE59 R5_11/5F | | 分子名称: | Kemp eliminase KE59 R5_11/5F, SULFATE ION | | 著者 | Khersonsky, O, Kiss, G, Roethlisberger, D, Dym, O, Albeck, S, Houk, K.N, Baker, D, Tawfik, D.S, Israel Structural Proteomics Center (ISPC) | | 登録日 | 2011-12-06 | | 公開日 | 2012-06-06 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.41 Å) | | 主引用文献 | Bridging the gaps in design methodologies by evolutionary optimization of the stability and proficiency of designed Kemp eliminase KE59.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
7RY2
 
 | | mSandy2 | | 分子名称: | mSandy2 | | 著者 | Chica, R.A, Legault, S, Thompson, M.C. | | 登録日 | 2021-08-24 | | 公開日 | 2022-01-26 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Generation of bright monomeric red fluorescent proteins via computational design of enhanced chromophore packing.
Chem Sci, 13, 2022
|
|
4PGL
 
 | | Crystal structure of engineered D-tagatose 3-epimerase PcDTE-ILS6 | | 分子名称: | D-tagatose 3-epimerase, L-sorbose, L-tagatose, ... | | 著者 | Hee, C.S, Bosshart, A, Schirmer, T. | | 登録日 | 2014-05-02 | | 公開日 | 2014-10-22 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Directed Divergent Evolution of a Thermostable D-Tagatose Epimerase towards Improved Activity for Two Hexose Substrates.
Chembiochem, 16, 2015
|
|
4Q7I
 
 | | Crystal structure of engineered thermostable D-tagatose 3-epimerase PcDTE-Var8 | | 分子名称: | D-tagatose 3-epimerase, GLYCEROL, IMIDAZOLE, ... | | 著者 | Hee, C.S, Bosshart, A, Schirmer, T. | | 登録日 | 2014-04-25 | | 公開日 | 2014-10-22 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Directed Divergent Evolution of a Thermostable D-Tagatose Epimerase towards Improved Activity for Two Hexose Substrates.
Chembiochem, 16, 2015
|
|
4PFH
 
 | | Crystal structure of engineered D-tagatose 3-epimerase PcDTE-IDF8 | | 分子名称: | D-fructose, D-psicose, D-tagatose 3-epimerase, ... | | 著者 | Hee, C.S, Bosshart, A, Schirmer, T. | | 登録日 | 2014-04-29 | | 公開日 | 2014-10-22 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Directed Divergent Evolution of a Thermostable D-Tagatose Epimerase towards Improved Activity for Two Hexose Substrates.
Chembiochem, 16, 2015
|
|
3FCZ
 
 | | Adaptive protein evolution grants organismal fitness by improving catalysis and flexibility | | 分子名称: | Beta-lactamase 2, ZINC ION | | 著者 | Tomatis, P, Fabiane, S, Simona, F, Carloni, P, Sutton, B, Vila, A. | | 登録日 | 2008-11-24 | | 公開日 | 2008-12-09 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.804 Å) | | 主引用文献 | Adaptive protein evolution grants organismal fitness by improving catalysis and flexibility.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
