5VNS
 
 | | M.tb Antigen 85C Acyl-Enzyme Intermediate with Tetrahydrolipstatin | | 分子名称: | (2S,3S,5S)-5-[(N-FORMYL-L-LEUCYL)OXY]-2-HEXYL-3-HYDROXYHEXADECANOIC ACID, (4S)-2-METHYL-2,4-PENTANEDIOL, Diacylglycerol acyltransferase/mycolyltransferase Ag85C, ... | | 著者 | Goins, C.M, Ronning, D.R. | | 登録日 | 2017-05-01 | | 公開日 | 2018-01-24 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Mycolyltransferase fromMycobacterium tuberculosisin covalent complex with tetrahydrolipstatin provides insights into antigen 85 catalysis.
J. Biol. Chem., 293, 2018
|
|
5VM7
 
 | | Pseudo-atomic model of the MurA-A2 complex | | 分子名称: | Maturation protein A2, UDP-N-acetylglucosamine 1-carboxyvinyltransferase | | 著者 | Cui, Z, Zhang, J. | | 登録日 | 2017-04-26 | | 公開日 | 2017-10-18 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (5.7 Å) | | 主引用文献 | Structures of Q beta virions, virus-like particles, and the Q beta-MurA complex reveal internal coat proteins and the mechanism of host lysis.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
8VJD
 
 | | Human R14A Pin1 covalently bound to inhibitor 158F10 in P21 21 21 space group | | 分子名称: | 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, Inhibitor 158F10, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1, ... | | 著者 | Rodriguez, I, Blaha, G. | | 登録日 | 2024-01-06 | | 公開日 | 2024-11-06 | | 最終更新日 | 2025-03-05 | | 実験手法 | X-RAY DIFFRACTION (1.57 Å) | | 主引用文献 | Targeted degradation of Pin1 by protein-destabilizing compounds.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8VPQ
 
 | | The structure of LSD1-CoREST-HDAC1 in complex with KBTBD4IPR310delinsTTYML | | 分子名称: | Histone deacetylase 1, INOSITOL HEXAKISPHOSPHATE, Isoform 2 of Kelch repeat and BTB domain-containing protein 4, ... | | 著者 | Xie, X, Liau, B, Zheng, N. | | 登録日 | 2024-01-16 | | 公開日 | 2024-11-27 | | 最終更新日 | 2025-06-11 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Converging mechanism of UM171 and KBTBD4 neomorphic cancer mutations.
Nature, 639, 2025
|
|
8VRT
 
 | | The structure of LSD1-CoREST-HDAC1 in complex with KBTBD4R313PRR mutant | | 分子名称: | Histone deacetylase 1, INOSITOL HEXAKISPHOSPHATE, Kelch repeat and BTB domain-containing protein 4, ... | | 著者 | Xie, X, Liau, B, Zheng, N. | | 登録日 | 2024-01-22 | | 公開日 | 2024-11-27 | | 最終更新日 | 2025-06-11 | | 実験手法 | ELECTRON MICROSCOPY (3.42 Å) | | 主引用文献 | Converging mechanism of UM171 and KBTBD4 neomorphic cancer mutations.
Nature, 639, 2025
|
|
5II7
 
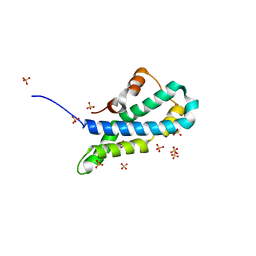 | | In-house sulfur-SAD structure of orthorhombic red abalone lysin at 1.66 A resolution | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Egg-lysin, SULFATE ION | | 著者 | Sadat Al-Hosseini, H, Raj, I, Nishimura, K, Jovine, L. | | 登録日 | 2016-03-01 | | 公開日 | 2017-06-14 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.66 Å) | | 主引用文献 | Structural Basis of Egg Coat-Sperm Recognition at Fertilization.
Cell, 169, 2017
|
|
2XCD
 
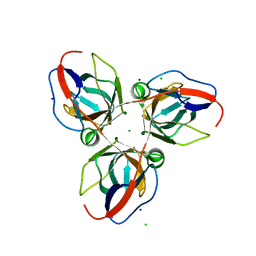 | | Structure of YncF,the genomic dUTPase from Bacillus subtilis | | 分子名称: | CHLORIDE ION, GLYCEROL, MAGNESIUM ION, ... | | 著者 | Garcia, J, Burchell, L, Takezawa, M, Rzechorzek, N.J, Fogg, M, Wilson, K.S. | | 登録日 | 2010-04-22 | | 公開日 | 2010-08-11 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.84 Å) | | 主引用文献 | The Structure of the Genomic Bacillus Subtilis Dutpase: Novel Features in the Phe-Lid.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
1KSD
 
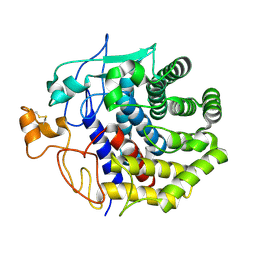 | | The structure of Endoglucanase from termite, Nasutitermes takasagoensis, at pH 6.5. | | 分子名称: | CALCIUM ION, Endo-b-1,4-glucanase | | 著者 | Khademi, S, Guarino, L.A, Watanabe, H, Tokuda, G, Meyer, E.F. | | 登録日 | 2002-01-12 | | 公開日 | 2003-01-21 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Structure of an endoglucanase from termite, Nasutitermes takasagoensis.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
4DMI
 
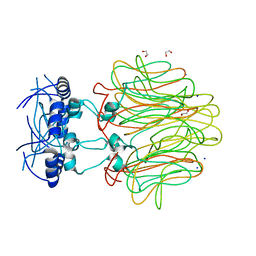 | | Crystal Structure of a Pentameric Capsid Protein Isolated from Metagenomic Phage Sequences (CASP) | | 分子名称: | 1,2-ETHANEDIOL, Capsid Protein, SODIUM ION | | 著者 | Craig, T.K, Abendroth, J, Lorimer, D, Burgin Jr, A.B, Segall, A, Rohwer, F. | | 登録日 | 2012-02-07 | | 公開日 | 2012-08-29 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Crystal Structure of a Pentameric Capsid Protein Isolated from Metagenomic Phage Sequences
To be Published
|
|
4M5A
 
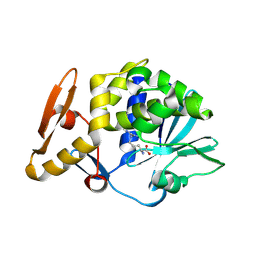 | | Crystal structure of the complex of Ribosome inactivating protein from Momordica balsamina inhibited by asymmetric dimethyl arginine at 1.70 A resolution | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, NG,NG-DIMETHYL-L-ARGININE, rRNA N-glycosidase | | 著者 | Yamini, S, Pandey, S, Kushwaha, G.S, Sinha, M, Bhushan, A, Kaur, P, Sharma, S, Singh, T.P. | | 登録日 | 2013-08-08 | | 公開日 | 2013-08-28 | | 最終更新日 | 2024-11-20 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Crystal structure of the complex of Ribosome inactivating protein from Momordica balsamina inhibited by asymmetric dimethyl arginine at 1.70 A resolution
To be Published
|
|
2XN4
 
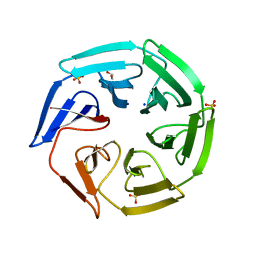 | | Crystal structure of the kelch domain of human KLHL2 (Mayven) | | 分子名称: | 1,2-ETHANEDIOL, KELCH-LIKE PROTEIN 2, SODIUM ION, ... | | 著者 | Canning, P, Hozjan, V, Cooper, C.D.O, Ayinampudi, V, Vollmar, M, Pike, A.C.W, von Delft, F, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A.N. | | 登録日 | 2010-07-30 | | 公開日 | 2010-08-18 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.99 Å) | | 主引用文献 | Structural Basis for Cul3 Assembly with the Btb-Kelch Family of E3 Ubiquitin Ligases.
J.Biol.Chem., 288, 2013
|
|
1KVL
 
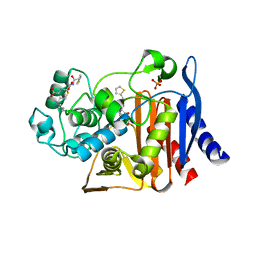 | | X-ray Crystal Structure of AmpC S64G Mutant beta-Lactamase in Complex with Substrate and Product Forms of Cephalothin | | 分子名称: | 2-[CARBOXY-(2-THIOPHEN-2-YL-ACETYLAMINO)-METHYL]-5-METHYL-3,6-DIHYDRO-2H-[1,3]THIAZINE-4-CARBOXYLIC ACID, 2-[CARBOXY-(2-THIOPHEN-2-YL-ACETYLAMINO)-METHYL]-5-METHYLENE-5,6-DIHYDRO-2H-[1,3]THIAZINE-4-CARBOXYLIC ACID, Beta-lactamase, ... | | 著者 | Beadle, B.M, Trehan, I, Focia, P.J, Shoichet, B.K. | | 登録日 | 2002-01-27 | | 公開日 | 2002-03-13 | | 最終更新日 | 2024-11-20 | | 実験手法 | X-RAY DIFFRACTION (1.53 Å) | | 主引用文献 | Structural milestones in the reaction pathway of an amide hydrolase: substrate, acyl, and product complexes of cephalothin with AmpC beta-lactamase.
Structure, 10, 2002
|
|
8VCK
 
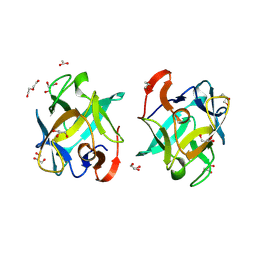 | | Galactose-binding lectin from Mytilus californianus, Isoform 1 (rMcL-1) | | 分子名称: | ACETATE ION, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | 著者 | Hernandez-Santoyo, A, Loera-Rubalcava, J. | | 登録日 | 2023-12-14 | | 公開日 | 2024-12-18 | | 最終更新日 | 2025-05-21 | | 実験手法 | X-RAY DIFFRACTION (1.49 Å) | | 主引用文献 | A mytilectin from Mytilus californianus: Study of its unique galactoside interactions, oligomerization patterns, and antifungal activity.
Int.J.Biol.Macromol., 308, 2025
|
|
9CP7
 
 | |
2XPC
 
 | | Second-generation sulfonamide inhibitors of MurD: Activity optimisation with conformationally rigid analogues of D-glutamic acid | | 分子名称: | (1R,3R,4S)-4-[({6-[(4-CYANO-2-FLUOROBENZYL)OXY]NAPHTHALEN-2-YL}SULFONYL)AMINO]CYCLOHEXANE-1,3-DICARBOXYLIC ACID, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | 著者 | Sosic, I, Barreteau, H, Simcic, M, Sink, R, Cesar, J, Golic-Grdadolnik, S, Contreras-Martel, C, Dessen, A, Amoroso, A, Joris, B, Blanot, D, Gobec, S. | | 登録日 | 2010-08-26 | | 公開日 | 2011-05-18 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.49 Å) | | 主引用文献 | Second-Generation Sulfonamide Inhibitors of D- Glutamic Acid-Adding Enzyme: Activity Optimisation with Conformationally Rigid Analogues of D- Glutamic Acid.
Eur.J.Med.Chem, 46, 2011
|
|
7M4H
 
 | | DNA Polymerase Lambda, dCTP:At Mn2+ Reaction State Ternary Complex, 225 min | | 分子名称: | 1,2-ETHANEDIOL, 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, DNA (5'-D(*CP*AP*GP*TP*AP*CP*C)-3'), ... | | 著者 | Jamsen, J.A, Wilson, S.H. | | 登録日 | 2021-03-21 | | 公開日 | 2022-07-06 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.923 Å) | | 主引用文献 | Watching right and wrong nucleotide insertion captures hidden polymerase fidelity checkpoints.
Nat Commun, 13, 2022
|
|
7M4B
 
 | | DNA Polymerase Lambda, TTP:At Mn2+ Product State Ternary Complex, 60 min | | 分子名称: | 1,2-ETHANEDIOL, DNA (5'-D(*CP*AP*GP*TP*AP*CP*T)-3'), DNA (5'-D(*CP*GP*GP*CP*AP*GP*TP*AP*CP*TP*G)-3'), ... | | 著者 | Jamsen, J.A, Wilson, S.H. | | 登録日 | 2021-03-21 | | 公開日 | 2022-07-06 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.66 Å) | | 主引用文献 | Watching right and wrong nucleotide insertion captures hidden polymerase fidelity checkpoints.
Nat Commun, 13, 2022
|
|
7M47
 
 | | DNA Polymerase Lambda, TTP:At Mg2+ Product State Ternary Complex, 60 min | | 分子名称: | 1,2-ETHANEDIOL, DNA (5'-D(*CP*AP*GP*TP*AP*CP*T)-3'), DNA (5'-D(*CP*GP*GP*CP*AP*GP*TP*AP*CP*TP*G)-3'), ... | | 著者 | Jamsen, J.A, Wilson, S.H. | | 登録日 | 2021-03-21 | | 公開日 | 2022-07-06 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.648 Å) | | 主引用文献 | Watching right and wrong nucleotide insertion captures hidden polymerase fidelity checkpoints.
Nat Commun, 13, 2022
|
|
5VH6
 
 | | 2.6 Angstrom Resolution Crystal Structure of N-terminal Fragment (residues 1-406) of Elongation Factor G from Bacillus subtilis. | | 分子名称: | CHLORIDE ION, Elongation factor G | | 著者 | Minasov, G, Shuvalova, L, Dubrovska, I, Kiryukhina, O, Grimshaw, S, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2017-04-12 | | 公開日 | 2017-04-19 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.61 Å) | | 主引用文献 | 2.6 Angstrom Resolution Crystal Structure of N-terminal Fragment (residues 1-406) of Elongation Factor G from Bacillus subtilis.
To Be Published
|
|
7M4J
 
 | | DNA Polymerase Lambda, dCTP:At Mn2+ Product State Ternary Complex, 960 min | | 分子名称: | 1,2-ETHANEDIOL, DNA (5'-D(*CP*AP*GP*TP*AP*CP*C)-3'), DNA (5'-D(*CP*GP*GP*CP*AP*GP*TP*AP*CP*TP*G)-3'), ... | | 著者 | Jamsen, J.A, Wilson, S.H. | | 登録日 | 2021-03-21 | | 公開日 | 2022-07-06 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.377 Å) | | 主引用文献 | Watching right and wrong nucleotide insertion captures hidden polymerase fidelity checkpoints.
Nat Commun, 13, 2022
|
|
7M4A
 
 | | DNA Polymerase Lambda, TTP:At Mn2+ Product State Ternary Complex, 20 min | | 分子名称: | 1,2-ETHANEDIOL, DNA (5'-D(*CP*AP*GP*TP*AP*CP*T)-3'), DNA (5'-D(*CP*GP*GP*CP*AP*GP*TP*AP*CP*TP*G)-3'), ... | | 著者 | Jamsen, J.A, Wilson, S.H. | | 登録日 | 2021-03-21 | | 公開日 | 2022-07-06 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.868 Å) | | 主引用文献 | Watching right and wrong nucleotide insertion captures hidden polymerase fidelity checkpoints.
Nat Commun, 13, 2022
|
|
7M46
 
 | | DNA Polymerase Lambda, TTP:At Mg2+ Product State Ternary Complex, 5 min | | 分子名称: | 1,2-ETHANEDIOL, DNA (5'-D(*CP*AP*GP*TP*AP*CP*T)-3'), DNA (5'-D(*CP*GP*GP*CP*AP*GP*TP*AP*CP*TP*G)-3'), ... | | 著者 | Jamsen, J.A, Wilson, S.H. | | 登録日 | 2021-03-21 | | 公開日 | 2022-07-06 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.92 Å) | | 主引用文献 | Watching right and wrong nucleotide insertion captures hidden polymerase fidelity checkpoints.
Nat Commun, 13, 2022
|
|
7M44
 
 | | DNA Polymerase Lambda, TTP:At Mg2+ Reaction State Ternary Complex, 90 sec | | 分子名称: | 1,2-ETHANEDIOL, DNA (5'-D(*CP*AP*GP*TP*AP*CP*T)-3'), DNA (5'-D(*CP*GP*GP*CP*AP*GP*TP*AP*CP*TP*G)-3'), ... | | 著者 | Jamsen, J.A, Wilson, S.H. | | 登録日 | 2021-03-21 | | 公開日 | 2022-07-06 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.901 Å) | | 主引用文献 | Watching right and wrong nucleotide insertion captures hidden polymerase fidelity checkpoints.
Nat Commun, 13, 2022
|
|
7M4C
 
 | | DNA Polymerase Lambda, TTP:At Mn2+ Product State Ternary Complex, 960 min | | 分子名称: | 1,2-ETHANEDIOL, DNA (5'-D(*CP*AP*GP*TP*AP*CP*T)-3'), DNA (5'-D(*CP*GP*GP*CP*AP*GP*TP*AP*CP*TP*G)-3'), ... | | 著者 | Jamsen, J.A, Wilson, S.H. | | 登録日 | 2021-03-21 | | 公開日 | 2022-07-06 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Watching right and wrong nucleotide insertion captures hidden polymerase fidelity checkpoints.
Nat Commun, 13, 2022
|
|
7M45
 
 | | DNA Polymerase Lambda, TTP:At Mg2+ Reaction State Ternary Complex, 120 sec | | 分子名称: | 1,2-ETHANEDIOL, DNA (5'-D(*CP*AP*GP*TP*AP*CP*T)-3'), DNA (5'-D(*CP*GP*GP*CP*AP*GP*TP*AP*CP*TP*G)-3'), ... | | 著者 | Jamsen, J.A, Wilson, S.H. | | 登録日 | 2021-03-21 | | 公開日 | 2022-07-06 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.889 Å) | | 主引用文献 | Watching right and wrong nucleotide insertion captures hidden polymerase fidelity checkpoints.
Nat Commun, 13, 2022
|
|
