7OTZ
 
 | | HIV-1 REVERSE TRANSCRIPTASE COMPLEX WITH DNA AND INHIBITOR RMC-259 | | 分子名称: | (S)-2-(2-(6-amino-9H-purin-9-yl)ethoxy)-3-phosphonopropanoic acid, DNA (5'-D(*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*(MRG)P*CP*GP*CP*CP*(DDG))-3'), DNA (5'-D(P*GP*GP*TP*CP*GP*GP*CP*GP*CP*CP*CP*GP*AP*AP*CP*AP*GP*GP*GP*AP*CP*TP*G)-3'), ... | | 著者 | Martinez, S.E, Singh, A.K, Gu, W, Das, K. | | 登録日 | 2021-06-10 | | 公開日 | 2021-12-08 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Exploring the dNTP -binding site of HIV-1 reverse transcriptase for inhibitor design.
Eur.J.Med.Chem., 225, 2021
|
|
7OUT
 
 | | HIV-1 REVERSE TRANSCRIPTASE COMPLEX WITH DNA AND INHIBITOR RMC-264 | | 分子名称: | (S)-2-(3-(6-amino-9H-purin-9-yl)propoxy)-3-phosphonopropanoic acid, DNA (5'-D(*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*(MRG)P*CP*GP*CP*CP*(DDG))-3'), DNA (5'-D(P*GP*GP*TP*CP*GP*GP*CP*GP*CP*CP*CP*GP*AP*AP*CP*AP*GP*GP*GP*AP*CP*TP*G)-3'), ... | | 著者 | Martinez, S.E, Singh, A.K, Gu, W, Das, K. | | 登録日 | 2021-06-13 | | 公開日 | 2021-12-08 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Exploring the dNTP -binding site of HIV-1 reverse transcriptase for inhibitor design.
Eur.J.Med.Chem., 225, 2021
|
|
7OTK
 
 | | HIV-1 REVERSE TRANSCRIPTASE COMPLEX WITH DNA AND INHIBITOR RMC-233 | | 分子名称: | (2~{R})-2-[2-(6-aminopurin-9-yl)ethylamino]-3-phosphono-propanoic acid, DNA (5'-D(*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*(MRG)*CP*GP*CP*CP*(DDG))-3'), DNA (5'-D(P*GP*GP*TP*CP*GP*GP*CP*GP*CP*CP*CP*GP*AP*AP*CP*AP*GP*GP*GP*AP*CP*TP*G)-3'), ... | | 著者 | Martinez, S.E, Singh, A.K, Gu, W, Das, K. | | 登録日 | 2021-06-10 | | 公開日 | 2021-12-08 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.95 Å) | | 主引用文献 | Exploring the dNTP -binding site of HIV-1 reverse transcriptase for inhibitor design.
Eur.J.Med.Chem., 225, 2021
|
|
7OTA
 
 | | HIV-1 REVERSE TRANSCRIPTASE COMPLEX WITH DNA AND INHIBITOR RMC-230 | | 分子名称: | ((2-(6-amino-9H-purin-9-yl)ethyl)-L-seryl)phosphoramidic acid, DNA (5'-D(*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*(MRG)*CP*GP*CP*CP*(DDG))-3'), DNA (5'-D(P*GP*GP*TP*CP*GP*GP*CP*GP*CP*CP*CP*GP*AP*AP*CP*AP*GP*GP*GP*AP*CP*TP*G)-3'), ... | | 著者 | Martinez, S.E, Singh, A.K, Gu, W, Das, K. | | 登録日 | 2021-06-09 | | 公開日 | 2021-12-08 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Exploring the dNTP -binding site of HIV-1 reverse transcriptase for inhibitor design.
Eur.J.Med.Chem., 225, 2021
|
|
8FWF
 
 | | Crystal structure of Apo form Fab235 | | 分子名称: | 1,2-ETHANEDIOL, ACETATE ION, CHLORIDE ION, ... | | 著者 | Tan, K, Kim, M, Reinherz, E.L. | | 登録日 | 2023-01-21 | | 公開日 | 2023-10-11 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (1.94 Å) | | 主引用文献 | Inadequate structural constraint on Fab approach rather than paratope elicitation limits HIV-1 MPER vaccine utility.
Nat Commun, 14, 2023
|
|
6WBZ
 
 | | Structure of Human HDAC2 in complex with an ethyl ketone inhibitor containing a spiro-bicyclic group | | 分子名称: | (1S)-N-{(1S)-7,7-dihydroxy-1-[5-(2-methoxyquinolin-3-yl)-1H-imidazol-2-yl]nonyl}-6-ethyl-6-azaspiro[2.5]octane-1-carboxamide, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Klein, D.J, Yu, W. | | 登録日 | 2020-03-28 | | 公開日 | 2020-05-06 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.32 Å) | | 主引用文献 | Discovery of ethyl ketone-based HDACs 1, 2, and 3 selective inhibitors for HIV latency reactivation.
Bioorg.Med.Chem.Lett., 30, 2020
|
|
8ZU2
 
 | | Crystal Structure of Human Myt1 Kinase domain Bounded with compound 8g | | 分子名称: | 2-azanyl-5-[2-(1,4-diazepan-1-yl)pyridin-4-yl]-3-(2,6-dimethyl-3-oxidanyl-phenyl)benzamide, GLYCINE, Membrane-associated tyrosine- and threonine-specific cdc2-inhibitory kinase | | 著者 | Zhang, Z.M, Zhou, Z.Q. | | 登録日 | 2024-06-07 | | 公開日 | 2024-09-11 | | 最終更新日 | 2024-09-25 | | 実験手法 | X-RAY DIFFRACTION (1.79888582 Å) | | 主引用文献 | Structure-Based Drug Design of 2-Amino-[1,1'-biphenyl]-3-carboxamide Derivatives as Selective PKMYT1 Inhibitors for the Treatment of CCNE1 -Amplified Breast Cancer.
J.Med.Chem., 67, 2024
|
|
8ZOS
 
 | | Cryo-EM structure of pyraclostrobin-bound porcine bc1 complex | | 分子名称: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, CARDIOLIPIN, Complex III subunit 9, ... | | 著者 | Wang, Y.X, Sun, J.Y, Li, Z.W, Cui, G.R, Yang, G.F. | | 登録日 | 2024-05-29 | | 公開日 | 2024-12-25 | | 最終更新日 | 2025-07-23 | | 実験手法 | ELECTRON MICROSCOPY (2.37 Å) | | 主引用文献 | Cryo-EM Structures Reveal the Unique Binding Modes of Metyltetraprole in Yeast and Porcine Cytochrome bc 1 Complex Enabling Rational Design of Inhibitors.
J.Am.Chem.Soc., 146, 2024
|
|
7PDA
 
 | | Crystal structure of Phenazine 1-carboxylic acid decarboxylase from Mycobacterium fortuitum | | 分子名称: | 1-deoxy-5-O-phosphono-1-(3,3,4,5-tetramethyl-9,11-dioxo-2,3,8,9,10,11-hexahydro-7H-quinolino[1,8-fg]pteridin-12-ium-7-y l)-D-ribitol, MANGANESE (II) ION, SODIUM ION, ... | | 著者 | Gahloth, D, Leys, D. | | 登録日 | 2021-08-05 | | 公開日 | 2022-08-24 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.65 Å) | | 主引用文献 | Crystal structure of Phenazine 1-carboxylic acid decarboxylase from Mycobacterium fortuitum
To Be Published
|
|
6X1V
 
 | | Structure of pHis Fab (SC44-8) in complex with pHis mimetic peptide | | 分子名称: | 1,2-ETHANEDIOL, ACLYana-3-pTza peptide, SC44-8 Heavy chain, ... | | 著者 | Kalagiri, R, Stanfield, R, Wilson, I.A, Hunter, T. | | 登録日 | 2020-05-19 | | 公開日 | 2021-02-03 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (2.11 Å) | | 主引用文献 | Structural basis for differential recognition of phosphohistidine-containing peptides by 1-pHis and 3-pHis monoclonal antibodies.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
1FPK
 
 | | FRUCTOSE-1,6-BISPHOSPHATASE (D-FRUCTOSE-1,6-BISPHOSPHATE 1-PHOSPHOHYDROLASE) COMPLEXED WITH THALLIUM IONS (10 MM) | | 分子名称: | FRUCTOSE-1,6-BISPHOSPHATASE, THALLIUM (I) ION | | 著者 | Villeret, V, Lipscomb, W.N. | | 登録日 | 1995-06-02 | | 公開日 | 1996-06-20 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Crystallographic evidence for the action of potassium, thallium, and lithium ions on fructose-1,6-bisphosphatase.
Proc.Natl.Acad.Sci.USA, 92, 1995
|
|
1G42
 
 | | STRUCTURE OF 1,3,4,6-TETRACHLORO-1,4-CYCLOHEXADIENE HYDROLASE (LINB) FROM SPHINGOMONAS PAUCIMOBILIS COMPLEXED WITH 1,2-DICHLOROPROPANE | | 分子名称: | 1,2-DICHLORO-PROPANE, 1,3,4,6-TETRACHLORO-1,4-CYCLOHEXADIENE HYDROLASE, ACETATE ION, ... | | 著者 | Oakley, A.J, Prokop, Z, Bohac, M, Kmunicek, J, Jedlicka, T, Monincova, M, Kuta-Smatanova, I, Nagata, Y, Damborsky, J, Wilce, M.C.J. | | 登録日 | 2000-10-26 | | 公開日 | 2001-10-26 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Exploring the structure and activity of haloalkane dehalogenase from Sphingomonas paucimobilis UT26: evidence for product- and water-mediated inhibition.
Biochemistry, 41, 2002
|
|
6XHH
 
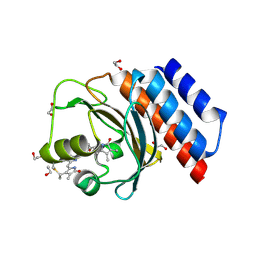 | | Far-red absorbing dark state of JSC1_58120g3 with bound 18-1, 18-2 dihydrobiliverdin IXa (DHBV), the native chromophore precursor | | 分子名称: | 1,2-ETHANEDIOL, JSC1_58120g3, mesobiliverdin IX(alpha) | | 著者 | Moreno, M.V, Rockwell, N.C, Fisher, A.J, Lagarias, J.C. | | 登録日 | 2020-06-18 | | 公開日 | 2020-10-28 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | A far-red cyanobacteriochrome lineage specific for verdins.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
8USY
 
 | | HIV-1 Integrase F185H N222K Complexed with Allosteric Inhibitor BI-D | | 分子名称: | (2S)-tert-butoxy[4-(3,4-dihydro-2H-chromen-6-yl)-2-methylquinolin-3-yl]ethanoic acid, Integrase | | 著者 | Montermoso, S, Gupta, K, Eilers, G, Bushman, F.D, Van Duyne, G.D. | | 登録日 | 2023-10-30 | | 公開日 | 2025-03-19 | | 最終更新日 | 2025-09-03 | | 実験手法 | X-RAY DIFFRACTION (4.4 Å) | | 主引用文献 | Structural Impact of Ex Vivo Resistance Mutations on HIV-1 Integrase Polymers Induced by Allosteric Inhibitors.
J.Mol.Biol., 437, 2025
|
|
8V0Z
 
 | | HIV-1 Integrase F185H W131C Complexed with Allosteric Inhibitor BI-D | | 分子名称: | (2S)-tert-butoxy[4-(3,4-dihydro-2H-chromen-6-yl)-2-methylquinolin-3-yl]ethanoic acid, Integrase | | 著者 | Montermoso, S, Gupta, K, Eilers, G, Bushman, F.D, Van Duyne, G.D. | | 登録日 | 2023-11-18 | | 公開日 | 2025-03-19 | | 最終更新日 | 2025-09-03 | | 実験手法 | X-RAY DIFFRACTION (4.56 Å) | | 主引用文献 | Structural Impact of Ex Vivo Resistance Mutations on HIV-1 Integrase Polymers Induced by Allosteric Inhibitors.
J.Mol.Biol., 437, 2025
|
|
1G4H
 
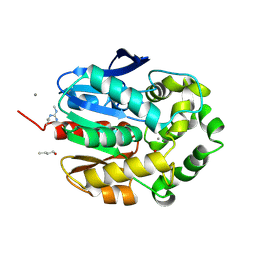 | | LINB COMPLEXED WITH BUTAN-1-OL | | 分子名称: | 1,3,4,6-TETRACHLORO-1,4-CYCLOHEXADIENE HYDROLASE, 1-BUTANOL, CALCIUM ION, ... | | 著者 | Oakley, A.J, Prokop, Z, Bohac, M, Kmunicek, J, Jedlicka, T, Monincova, M, Kuta-Smatanova, I, Nagata, Y, Damborsky, J, Wilce, M.C.J. | | 登録日 | 2000-10-27 | | 公開日 | 2001-10-27 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Exploring the structure and activity of haloalkane dehalogenase from Sphingomonas paucimobilis UT26: evidence for product- and water-mediated inhibition.
Biochemistry, 41, 2002
|
|
8BNQ
 
 | |
5WJA
 
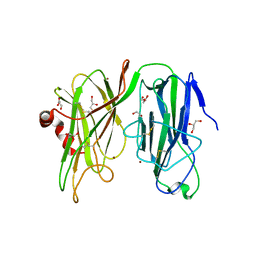 | | Crystal structure of H107A peptidylglycine alpha-hydroxylating monooxygenase (PHM) in complex with citrate | | 分子名称: | CITRATE ANION, COPPER (II) ION, GLYCEROL, ... | | 著者 | Maheshwari, S, Rudzka, K, Gabelli, S.B, Amzel, L.M. | | 登録日 | 2017-07-21 | | 公開日 | 2018-07-18 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Effects of copper occupancy on the conformational landscape of peptidylglycine alpha-hydroxylating monooxygenase.
Commun Biol, 1, 2018
|
|
8V38
 
 | |
8Q2E
 
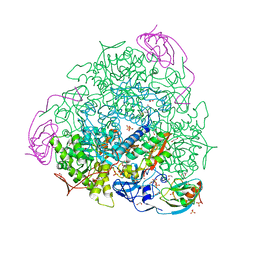 | | The 1.68-A X-ray crystal structure of Sporosarcina pasteurii urease inhibited by thiram and bound to dimethylditiocarbamate | | 分子名称: | 1,2-ETHANEDIOL, HYDROXIDE ION, NICKEL (II) ION, ... | | 著者 | Mazzei, L, Cianci, M, Ciurli, S. | | 登録日 | 2023-08-02 | | 公開日 | 2023-11-01 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.68 Å) | | 主引用文献 | Kinetic and structural details of urease inactivation by thiuram disulphides.
J.Inorg.Biochem., 250, 2023
|
|
6WEB
 
 | | Multi-Hit SFX using MHz XFEL sources | | 分子名称: | 1,2-ETHANEDIOL, ACETATE ION, CHLORIDE ION, ... | | 著者 | Holmes, S, Darmanin, C, Abbey, B. | | 登録日 | 2020-04-01 | | 公開日 | 2021-10-13 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Megahertz pulse trains enable multi-hit serial femtosecond crystallography experiments at X-ray free electron lasers.
Nat Commun, 13, 2022
|
|
6WEC
 
 | | Multi-Hit SFX using MHz XFEL sources | | 分子名称: | 1,2-ETHANEDIOL, ACETATE ION, CHLORIDE ION, ... | | 著者 | Holmes, S, Darmanin, C, Abbey, B. | | 登録日 | 2020-04-01 | | 公開日 | 2021-10-13 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Megahertz pulse trains enable multi-hit serial femtosecond crystallography experiments at X-ray free electron lasers.
Nat Commun, 13, 2022
|
|
6UZY
 
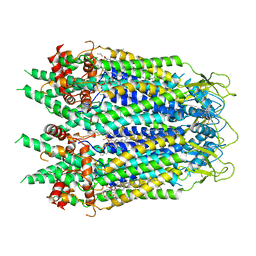 | | Cryo-EM structure of Xenopus tropicalis pannexin 1 | | 分子名称: | HEXADECANE, Pannexin, [(2~{R})-1-[2-azanylethoxy(oxidanyl)phosphoryl]oxy-3-hexadecanoyloxy-propan-2-yl] (~{Z})-octadec-9-enoate | | 著者 | Deng, Z, He, Z, Yuan, P. | | 登録日 | 2019-11-15 | | 公開日 | 2020-04-01 | | 最終更新日 | 2024-10-23 | | 実験手法 | ELECTRON MICROSCOPY (3.38 Å) | | 主引用文献 | Cryo-EM structures of the ATP release channel pannexin 1.
Nat.Struct.Mol.Biol., 27, 2020
|
|
7T7E
 
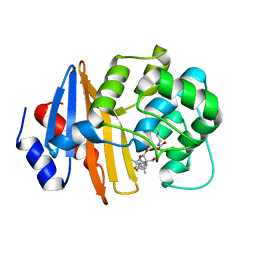 | | MA-1-206-OXA-23 3 minute complex | | 分子名称: | (2R,4S)-2-(1,3-dihydroxypropan-2-yl)-4-{[(3R,5R)-5-(dimethylcarbamoyl)pyrrolidin-3-yl]sulfanyl}-3,4-dihydro-2H-pyrrole-5-carboxylic acid, Beta-lactamase OXA-23 | | 著者 | Smith, C.A, Stewart, N.K, Vakulenko, S.B. | | 登録日 | 2021-12-15 | | 公開日 | 2022-05-18 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | C6 Hydroxymethyl-Substituted Carbapenem MA-1-206 Inhibits the Major Acinetobacter baumannii Carbapenemase OXA-23 by Impeding Deacylation.
Mbio, 13, 2022
|
|
7T7F
 
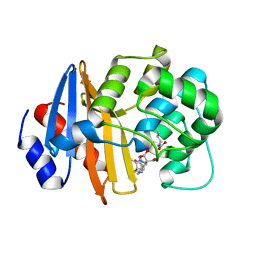 | | MA-1-206-OXA-23 25 minute complex | | 分子名称: | (2R,4S)-2-(1,3-dihydroxypropan-2-yl)-4-{[(3R,5R)-5-(dimethylcarbamoyl)pyrrolidin-3-yl]sulfanyl}-3,4-dihydro-2H-pyrrole-5-carboxylic acid, Beta-lactamase OXA-23 | | 著者 | Smith, C.A, Stewart, N.K, Vakulenko, S.B. | | 登録日 | 2021-12-15 | | 公開日 | 2022-05-18 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | C6 Hydroxymethyl-Substituted Carbapenem MA-1-206 Inhibits the Major Acinetobacter baumannii Carbapenemase OXA-23 by Impeding Deacylation.
Mbio, 13, 2022
|
|
