1E10
 
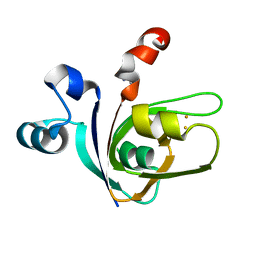 | | [2Fe-2S]-Ferredoxin from Halobacterium salinarum | | 分子名称: | ACETYL GROUP, FE2/S2 (INORGANIC) CLUSTER, FERREDOXIN | | 著者 | Marg, B.-L, Schweimer, K, Oesterhelt, D, Roesch, P, Sticht, H. | | 登録日 | 2000-04-12 | | 公開日 | 2001-04-09 | | 最終更新日 | 2011-07-13 | | 実験手法 | SOLUTION NMR | | 主引用文献 | A Two-Alpha-Helix Extra Domain Mediates the Halophilic Character of a Plant-Type Ferredoxin from Halophilic Archaea.
Biochemistry, 44, 2005
|
|
1NT5
 
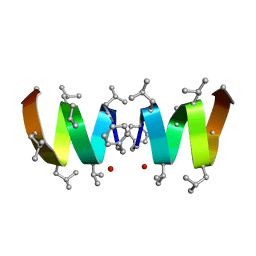 | | F1-Gramicidin A in Sodium Dodecyl Sulfate Micelles (NMR) | | 分子名称: | GRAMICIDIN A | | 著者 | Townsley, L.E, Fletcher, T.G, Hinton, J.F. | | 登録日 | 2003-01-28 | | 公開日 | 2003-02-11 | | 最終更新日 | 2023-11-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The Structure, Cation Binding, Transport, and Conductance of Gly15-Gramicidin a Incorporated Into Sds Micelles and Pc/Pg Vesicles.
Biochemistry, 42, 2003
|
|
6XUZ
 
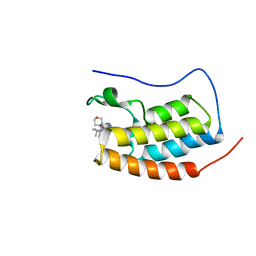 | | CRYSTAL STRUCTURE OF BRD4-BD1 WITH COMPOUND 4 | | 分子名称: | 6-[1-[(2~{S})-1-methoxypropan-2-yl]-6-[(3~{S})-3-methylmorpholin-4-yl]imidazo[4,5-c]pyridin-2-yl]-3-methyl-~{N}-propan-2-yl-[1,2,4]triazolo[4,3-a]pyrazin-8-amine, Bromodomain-containing protein 4 | | 著者 | Bader, G, Kessler, D, Wolkerstorfer, B. | | 登録日 | 2020-01-21 | | 公開日 | 2020-07-08 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.07 Å) | | 主引用文献 | PI by NMR: Probing CH-pi Interactions in Protein-Ligand Complexes by NMR Spectroscopy.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6XV3
 
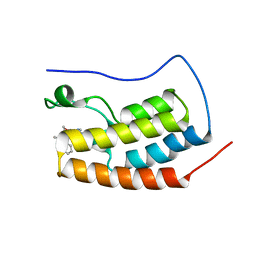 | | CRYSTAL STRUCTURE OF BRD4-BD1 WITH COMPOUND 3 | | 分子名称: | 3-methyl-6-[6-[(3~{S})-3-methylmorpholin-4-yl]-1-[(1~{S})-1-phenylethyl]imidazo[4,5-c]pyridin-2-yl]-~{N}-propan-2-yl-[1,2,4]triazolo[4,3-a]pyrazin-8-amine, Bromodomain-containing protein 4 | | 著者 | Bader, G, Kessler, D, Wolkerstorfer, B. | | 登録日 | 2020-01-21 | | 公開日 | 2020-07-08 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.47 Å) | | 主引用文献 | PI by NMR: Probing CH-pi Interactions in Protein-Ligand Complexes by NMR Spectroscopy.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
1F7X
 
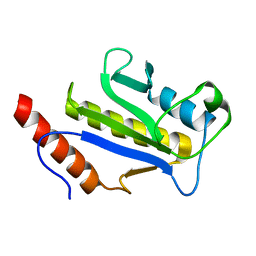 | | SOLUTION STRUCTURE OF C-TERMINAL DOMAIN ZIPA | | 分子名称: | CELL DIVISION PROTEIN ZIPA | | 著者 | Moy, F.J, Glasfeld, E, Mosyak, L, Powers, R. | | 登録日 | 2000-06-28 | | 公開日 | 2001-06-28 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of ZipA, a crucial component of Escherichia coli cell division.
Biochemistry, 39, 2000
|
|
6XVC
 
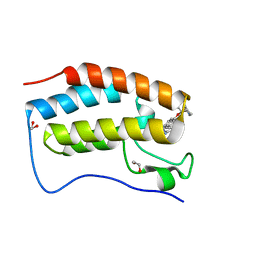 | | CRYSTAL STRUCTURE OF BRD4-BD1 WITH COMPOUND 1 | | 分子名称: | (4~{R})-4-[(1~{R})-1-[7-(3-methyl-[1,2,4]triazolo[4,3-a]pyridin-6-yl)quinolin-5-yl]oxyethyl]pyrrolidin-2-one, 1,2-ETHANEDIOL, Bromodomain-containing protein 4 | | 著者 | Bader, G, Kessler, D, Wolkerstorfer, B. | | 登録日 | 2020-01-21 | | 公開日 | 2020-07-08 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.098 Å) | | 主引用文献 | PI by NMR: Probing CH-pi Interactions in Protein-Ligand Complexes by NMR Spectroscopy.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6XV7
 
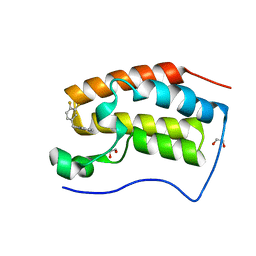 | | CRYSTAL STRUCTURE OF BRD4-BD1 WITH COMPOUND 2 | | 分子名称: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, ~{N}-[[3,4-bis(fluoranyl)phenyl]methyl]-~{N},3-dimethyl-[1,2,4]triazolo[4,3-b]pyridazin-6-amine | | 著者 | Bader, G, Kessler, D, Wolkerstorfer, B. | | 登録日 | 2020-01-21 | | 公開日 | 2020-07-08 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.668 Å) | | 主引用文献 | PI by NMR: Probing CH-pi Interactions in Protein-Ligand Complexes by NMR Spectroscopy.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6AX2
 
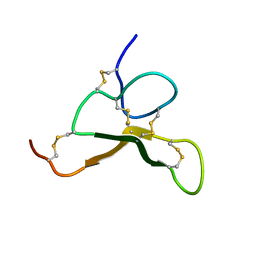 | |
6B9K
 
 | |
1AXL
 
 | | SOLUTION CONFORMATION OF THE (-)-TRANS-ANTI-[BP]DG ADDUCT OPPOSITE A DELETION SITE IN DNA DUPLEX D(CCATC-[BP]G-CTACC)D(GGTAG--GATGG), NMR, 6 STRUCTURES | | 分子名称: | 1,2,3-TRIHYDROXY-1,2,3,4-TETRAHYDROBENZO[A]PYRENE, DNA DUPLEX D(CCATC-[BP]G-CTACC)D(GGTAG--GATGG) | | 著者 | Feng, B, Gorin, A.A, Kolbanovskiy, A, Hingerty, B.E, Geacintov, N.E, Broyde, S, Patel, D.J. | | 登録日 | 1997-10-16 | | 公開日 | 1998-07-01 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution conformation of the (-)-trans-anti-[BP]dG adduct opposite a deletion site in a DNA duplex: intercalation of the covalently attached benzo[a]pyrene into the helix with base displacement of the modified deoxyguanosine into the minor groove.
Biochemistry, 36, 1997
|
|
6BHN
 
 | | Red Light-Absorbing State of NpR6012g4, a Red/Green Cyanobacteriochrome | | 分子名称: | Methyl-accepting chemotaxis sensory transducer with phytochrome sensor, PHYCOCYANOBILIN | | 著者 | Yu, Q, Lim, S, Rockwell, N.C, Martin, S.S, Lagarias, J.C, Ames, J.B. | | 登録日 | 2017-10-31 | | 公開日 | 2018-04-18 | | 最終更新日 | 2019-12-04 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Correlating structural and photochemical heterogeneity in cyanobacteriochrome NpR6012g4.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6BGH
 
 | |
1NT6
 
 | | F1-Gramicidin C In Sodium Dodecyl Sulfate Micelles (NMR) | | 分子名称: | GRAMICIDIN C | | 著者 | Townsley, L.E, Fletcher, T.G, Hinton, J.F. | | 登録日 | 2003-01-28 | | 公開日 | 2003-02-11 | | 最終更新日 | 2023-11-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The Structure, Cation Binding, Transport, and Conductance of Gly15-Gramicidin a Incorporated Into Sds Micelles and Pc/Pg Vesicles.
Biochemistry, 42, 2003
|
|
6BTV
 
 | | Solution NMR structures for CcoTx-II | | 分子名称: | Beta-theraphotoxin-Cm1b | | 著者 | Agwa, A.J, Schroeder, C.I. | | 登録日 | 2017-12-07 | | 公開日 | 2018-05-09 | | 最終更新日 | 2023-06-14 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Gating modifier toxins isolated from spider venom: Modulation of voltage-gated sodium channels and the role of lipid membranes.
J. Biol. Chem., 293, 2018
|
|
6BR0
 
 | | Solution NMR structure for CcoTx-I | | 分子名称: | Beta-theraphotoxin-Cm1a | | 著者 | Agwa, A.J, Schroeder, C.I. | | 登録日 | 2017-11-29 | | 公開日 | 2018-05-16 | | 最終更新日 | 2023-06-14 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Gating modifier toxins isolated from spider venom: Modulation of voltage-gated sodium channels and the role of lipid membranes.
J. Biol. Chem., 293, 2018
|
|
6BGG
 
 | | Solution NMR structures of the BRD3 ET domain in complex with a CHD4 peptide | | 分子名称: | Bromodomain-containing protein 3, CHD4 | | 著者 | Wai, D.C.C, Szyszka, T.N, Campbell, A.E, Kwong, C, Wilkinson-White, L, Silva, A.P.G, Low, J.K.K, Kwan, A.H, Gamsjaeger, R, Lu, B, Vakoc, C.R, Blobel, G.A, Mackay, J.P. | | 登録日 | 2017-10-28 | | 公開日 | 2018-03-21 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The BRD3 ET domain recognizes a short peptide motif through a mechanism that is conserved across chromatin remodelers and transcriptional regulators.
J. Biol. Chem., 293, 2018
|
|
1BJ6
 
 | | 1H NMR OF (12-53) NCP7/D(ACGCC) COMPLEX, 10 STRUCTURES | | 分子名称: | DNA (5'-D(*AP*CP*GP*CP*C)-3'), NUCLEOCAPSID PROTEIN 7, ZINC ION | | 著者 | Demene, H, Morellet, N, Teilleux, V, Huynh-Dinh, T, De Rocquigny, H, Fournie-Zaluski, M.C, Roques, B.P. | | 登録日 | 1998-07-03 | | 公開日 | 1999-02-02 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structure of the complex between the HIV-1 nucleocapsid protein NCp7 and the single-stranded pentanucleotide d(ACGCC).
J.Mol.Biol., 283, 1998
|
|
1BXP
 
 | | SOLUTION NMR STRUCTURE OF THE COMPLEX OF ALPHA-BUNGAROTOXIN WITH A LIBRARY DERIVED PEPTIDE, 20 STRUCTURES | | 分子名称: | ALPHA-BUNGAROTOXIN, PEPTIDE MET-ARG-TYR-TYR-GLU-SER-SER-LEU-LYS-SER-TYR-PRO-ASP | | 著者 | Scherf, T, Balass, M, Fuchs, S, Katchalski-Katzir, E, Anglister, J. | | 登録日 | 1998-08-23 | | 公開日 | 1999-01-27 | | 最終更新日 | 2022-02-16 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Three-dimensional solution structure of the complex of alpha-bungarotoxin with a library-derived peptide.
Proc.Natl.Acad.Sci.USA, 94, 1997
|
|
6C0A
 
 | | Actinin-1 EF-Hand bound to the Cav1.2 IQ Motif | | 分子名称: | Alpha-actinin-1, Voltage-dependent L-type calcium channel subunit alpha-1C | | 著者 | Turner, M.L, Ames, J.B. | | 登録日 | 2017-12-28 | | 公開日 | 2019-01-16 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural Basis of the Localization and Activation of Neuronal L-type Ca2+ Channels by a-Actinin1 and Calmodulin
To be Published
|
|
6BHO
 
 | | Green Light-Absorbing State of NpR6012g4, a Red/Green Cyanobacteriochrome | | 分子名称: | Methyl-accepting chemotaxis sensory transducer with phytochrome sensor, PHYCOCYANOBILIN | | 著者 | Lim, S, Yu, Q, Rockwell, N.C, Martin, S.S, Lagarias, J.C, Ames, J.B. | | 登録日 | 2017-10-31 | | 公開日 | 2018-04-18 | | 最終更新日 | 2019-12-04 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Correlating structural and photochemical heterogeneity in cyanobacteriochrome NpR6012g4.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
1CXR
 
 | |
1CYE
 
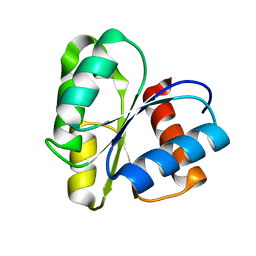 | | THREE DIMENSIONAL STRUCTURE OF CHEMOTACTIC CHE Y PROTEIN IN AQUEOUS SOLUTION BY NUCLEAR MAGNETIC RESONANCE METHODS | | 分子名称: | CHEY | | 著者 | Santoro, J, Bruix, M, Pascual, J, Lopez, E, Serrano, L, Rico, M. | | 登録日 | 1994-10-21 | | 公開日 | 1995-02-07 | | 最終更新日 | 2024-04-10 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Three-dimensional structure of chemotactic Che Y protein in aqueous solution by nuclear magnetic resonance methods.
J.Mol.Biol., 247, 1995
|
|
1QN1
 
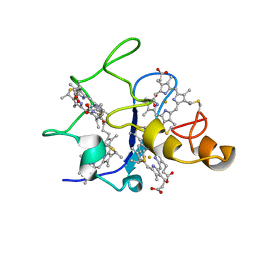 | | SOLUTION STRUCTURE OF DESULFOVIBRIO GIGAS FERRICYTOCHROME C3, NMR, 15 STRUCTURES | | 分子名称: | CYTOCHROME C3, HEME C | | 著者 | Brennan, L, Messias, A.C, Legall, J, Turner, D.L, Xavier, A.V. | | 登録日 | 1999-10-11 | | 公開日 | 2000-10-12 | | 最終更新日 | 2019-11-06 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural Basis for the Network of Functional Cooperativities in Cytochromes C3 from Desulfovibrio Gigas: Solution Structures of the Oxidised and Reduced States
J.Mol.Biol., 298, 2000
|
|
1N3G
 
 | |
1MWB
 
 | | Solution structure of the recombinant hemoglobin from the cyanobacterium Synechocystis sp. PCC 6803 in its hemichrome state | | 分子名称: | Cyanoglobin, PROTOPORPHYRIN IX CONTAINING FE | | 著者 | Falzone, C.J, Vu, B.C, Scott, N.L, Lecomte, J.T. | | 登録日 | 2002-09-27 | | 公開日 | 2002-12-04 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The Solution Structure of the Recombinant Hemoglobin from the Cyanobacterium Synechocystis sp. PCC 6803 in its Hemichrome State
J.MOL.BIOL., 324, 2002
|
|
