7MMI
 
 | | Crystal structure of HCV NS3/4A D168A protease in complex with NR02-23 | | 分子名称: | (2S)-1,1,1-trifluoropropan-2-yl {(2R,4S,6S,12Z,13aS,14aR,16aS)-2-[(7-methoxy-3-methylquinoxalin-2-yl)oxy]-14a-[(1-methylcyclopropane-1-sulfonyl)carbamoyl]-5,16-dioxo-1,2,3,5,6,7,8,9,10,11,13a,14,14a,15,16,16a-hexadecahydrocyclopropa[e]pyrrolo[1,2-a][1,4]diazacyclopentadecin-6-yl}carbamate, NS3/4A protease, ZINC ION | | 著者 | Zephyr, J, Schiffer, C.A. | | 登録日 | 2021-04-29 | | 公開日 | 2022-03-09 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Deciphering the Molecular Mechanism of HCV Protease Inhibitor Fluorination as a General Approach to Avoid Drug Resistance.
J.Mol.Biol., 434, 2022
|
|
7MM5
 
 | | Crystal structure of HCV NS3/4A protease in complex with NR02-60 | | 分子名称: | (1R)-1-cyclopentyl-2,2,2-trifluoroethyl {(2R,4R,6S,12Z,13aS,14aR,16aS)-2-[(7-methoxy-3-methylquinoxalin-2-yl)oxy]-14a-[(1-methylcyclopropane-1-sulfonyl)carbamoyl]-5,16-dioxo-1,2,3,5,6,7,8,9,10,11,13a,14,14a,15,16,16a-hexadecahydrocyclopropa[e]pyrrolo[1,2-a][1,4]diazacyclopentadecin-6-yl}carbamate, 1,2-ETHANEDIOL, NS3/4a protease, ... | | 著者 | Zephyr, J, Schiffer, C.A. | | 登録日 | 2021-04-29 | | 公開日 | 2022-03-09 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.86 Å) | | 主引用文献 | Deciphering the Molecular Mechanism of HCV Protease Inhibitor Fluorination as a General Approach to Avoid Drug Resistance.
J.Mol.Biol., 434, 2022
|
|
7MMG
 
 | | Crystal structure of HCV NS3/4A D168A protease in complex with NR02-58 | | 分子名称: | 1-(trifluoromethyl)cyclobutyl {(2R,4S,6S,12Z,13aS,14aR,16aS)-2-[(7-methoxy-3-methylquinoxalin-2-yl)oxy]-14a-[(1-methylcyclopropane-1-sulfonyl)carbamoyl]-5,16-dioxo-1,2,3,5,6,7,8,9,10,11,13a,14,14a,15,16,16a-hexadecahydrocyclopropa[e]pyrrolo[1,2-a][1,4]diazacyclopentadecin-6-yl}carbamate, ARGININE, NS3/4A protease, ... | | 著者 | Zephyr, J, Schiffer, C.A. | | 登録日 | 2021-04-29 | | 公開日 | 2022-03-09 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Deciphering the Molecular Mechanism of HCV Protease Inhibitor Fluorination as a General Approach to Avoid Drug Resistance.
J.Mol.Biol., 434, 2022
|
|
7MML
 
 | | Crystal structure of HCV NS3/4A D168A protease in complex with NR01-145 | | 分子名称: | (2R)-1,1,1-trifluoropropan-2-yl {(2R,4R,6S,12Z,13aS,14aR,16aS)-2-[(7-methoxy-3-methylquinoxalin-2-yl)oxy]-14a-[(1-methylcyclopropane-1-sulfonyl)carbamoyl]-5,16-dioxo-1,2,3,5,6,7,8,9,10,11,13a,14,14a,15,16,16a-hexadecahydrocyclopropa[e]pyrrolo[1,2-a][1,4]diazacyclopentadecin-6-yl}carbamate, NS3 protease, SULFATE ION, ... | | 著者 | Zephyr, J, Schiffer, C.A. | | 登録日 | 2021-04-29 | | 公開日 | 2022-03-16 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.701 Å) | | 主引用文献 | Deciphering the Molecular Mechanism of HCV Protease Inhibitor Fluorination as a General Approach to Avoid Drug Resistance.
J.Mol.Biol., 434, 2022
|
|
7MMH
 
 | |
7MMK
 
 | | Crystal structure of HCV NS3/4A D168A protease in complex with NR01-149 | | 分子名称: | 1,2-ETHANEDIOL, 3,3-difluorocyclobutyl {(2R,4S,6S,12Z,13aS,14aR,16aS)-2-[(7-methoxy-3-methylquinoxalin-2-yl)oxy]-14a-[(1-methylcyclopropane-1-sulfonyl)carbamoyl]-5,16-dioxo-1,2,3,5,6,7,8,9,10,11,13a,14,14a,15,16,16a-hexadecahydrocyclopropa[e]pyrrolo[1,2-a][1,4]diazacyclopentadecin-6-yl}carbamate, NS3 protease, ... | | 著者 | Zephyr, J, Schiffer, C.A. | | 登録日 | 2021-04-29 | | 公開日 | 2022-03-16 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.89 Å) | | 主引用文献 | Deciphering the Molecular Mechanism of HCV Protease Inhibitor Fluorination as a General Approach to Avoid Drug Resistance.
J.Mol.Biol., 434, 2022
|
|
7MM9
 
 | | Crystal structure of HCV NS3/4A protease in complex with NR01-149 | | 分子名称: | 1,2-ETHANEDIOL, 3,3-difluorocyclobutyl {(2R,4S,6S,12Z,13aS,14aR,16aS)-2-[(7-methoxy-3-methylquinoxalin-2-yl)oxy]-14a-[(1-methylcyclopropane-1-sulfonyl)carbamoyl]-5,16-dioxo-1,2,3,5,6,7,8,9,10,11,13a,14,14a,15,16,16a-hexadecahydrocyclopropa[e]pyrrolo[1,2-a][1,4]diazacyclopentadecin-6-yl}carbamate, NS3 protease, ... | | 著者 | Zephyr, J, Schiffer, C.A. | | 登録日 | 2021-04-29 | | 公開日 | 2022-03-16 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.11 Å) | | 主引用文献 | Deciphering the Molecular Mechanism of HCV Protease Inhibitor Fluorination as a General Approach to Avoid Drug Resistance.
J.Mol.Biol., 434, 2022
|
|
7MMA
 
 | | Crystal structure of HCV NS3/4A protease in complex with NR01-145 | | 分子名称: | (2R)-1,1,1-trifluoropropan-2-yl {(2R,4R,6S,12Z,13aS,14aR,16aS)-2-[(7-methoxy-3-methylquinoxalin-2-yl)oxy]-14a-[(1-methylcyclopropane-1-sulfonyl)carbamoyl]-5,16-dioxo-1,2,3,5,6,7,8,9,10,11,13a,14,14a,15,16,16a-hexadecahydrocyclopropa[e]pyrrolo[1,2-a][1,4]diazacyclopentadecin-6-yl}carbamate, 1,2-ETHANEDIOL, NS3 protease, ... | | 著者 | Zephyr, J, Schiffer, C.A. | | 登録日 | 2021-04-29 | | 公開日 | 2022-03-16 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Deciphering the Molecular Mechanism of HCV Protease Inhibitor Fluorination as a General Approach to Avoid Drug Resistance.
J.Mol.Biol., 434, 2022
|
|
7MM3
 
 | |
7MMB
 
 | |
7MMC
 
 | | Crystal structure of HCV NS3/4A D168A protease in complex with NR01-115 | | 分子名称: | (1-methylcyclopropyl)methyl {(2R,4S,6S,12Z,13aS,14aR,16aS)-2-[(7-methoxy-3-methylquinoxalin-2-yl)oxy]-14a-[(1-methylcyclopropane-1-sulfonyl)carbamoyl]-5,16-dioxo-1,2,3,5,6,7,8,9,10,11,13a,14,14a,15,16,16a-hexadecahydrocyclopropa[e]pyrrolo[1,2-a][1,4]diazacyclopentadecin-6-yl}carbamate, 1,2-ETHANEDIOL, NS3 protease, ... | | 著者 | Zephyr, J, Schiffer, C.A. | | 登録日 | 2021-04-29 | | 公開日 | 2022-03-16 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.001 Å) | | 主引用文献 | Deciphering the Molecular Mechanism of HCV Protease Inhibitor Fluorination as a General Approach to Avoid Drug Resistance.
J.Mol.Biol., 434, 2022
|
|
7MM4
 
 | | Crystal structure of HCV NS3/4A protease in complex with NR01-115 | | 分子名称: | (1-methylcyclopropyl)methyl {(2R,4S,6S,12Z,13aS,14aR,16aS)-2-[(7-methoxy-3-methylquinoxalin-2-yl)oxy]-14a-[(1-methylcyclopropane-1-sulfonyl)carbamoyl]-5,16-dioxo-1,2,3,5,6,7,8,9,10,11,13a,14,14a,15,16,16a-hexadecahydrocyclopropa[e]pyrrolo[1,2-a][1,4]diazacyclopentadecin-6-yl}carbamate, 1,2-ETHANEDIOL, NS3 protease, ... | | 著者 | Zephyr, J, Schiffer, C.A. | | 登録日 | 2021-04-29 | | 公開日 | 2022-03-16 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.89 Å) | | 主引用文献 | Deciphering the Molecular Mechanism of HCV Protease Inhibitor Fluorination as a General Approach to Avoid Drug Resistance.
J.Mol.Biol., 434, 2022
|
|
4AX3
 
 | | Structure of three-domain heme-Cu nitrite reductase from Ralstonia pickettii at 1.6 A resolution | | 分子名称: | COPPER (II) ION, COPPER-CONTAINING NITRITE REDUCTASE, HEME C | | 著者 | Antonyuk, S.V, Cong, H, Eady, R.R, Hasnain, S.S. | | 登録日 | 2012-06-07 | | 公開日 | 2013-03-27 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Structures of protein-protein complexes involved in electron transfer.
Nature, 496, 2013
|
|
7Y5B
 
 | | Cryo-EM structure of F-ATP synthase from Mycolicibacterium smegmatis (rotational state 1) | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase epsilon chain, ... | | 著者 | Saw, W.-G, Wong, C.F, Grueber, G. | | 登録日 | 2022-06-16 | | 公開日 | 2022-11-23 | | 最終更新日 | 2024-07-03 | | 実験手法 | ELECTRON MICROSCOPY (4.4 Å) | | 主引用文献 | Structural Elements Involved in ATP Hydrolysis Inhibition and ATP Synthesis of Tuberculosis and Nontuberculous Mycobacterial F-ATP Synthase Decipher New Targets for Inhibitors.
Antimicrob.Agents Chemother., 66, 2022
|
|
4EHZ
 
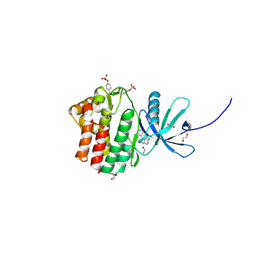 | | The Jak1 kinase domain in complex with inhibitor | | 分子名称: | 1,2-ETHANEDIOL, 2-methyl-1-(piperidin-4-yl)-1,6-dihydroimidazo[4,5-d]pyrrolo[2,3-b]pyridine, Tyrosine-protein kinase JAK1 | | 著者 | Lupardus, P.J, Steffek, M. | | 登録日 | 2012-04-04 | | 公開日 | 2012-07-04 | | 最終更新日 | 2013-01-23 | | 実験手法 | X-RAY DIFFRACTION (2.174 Å) | | 主引用文献 | Discovery and optimization of C-2 methyl imidazopyrrolopyridines as potent and orally bioavailable JAK1 inhibitors with selectivity over JAK2.
J.Med.Chem., 55, 2012
|
|
3ZKP
 
 | |
1QBQ
 
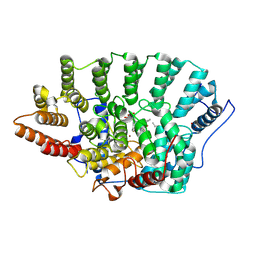 | | STRUCTURE OF RAT FARNESYL PROTEIN TRANSFERASE COMPLEXED WITH A CVIM PEPTIDE AND ALPHA-HYDROXYFARNESYLPHOSPHONIC ACID. | | 分子名称: | ACETATE ION, ACETYL-CYS-VAL-ILE-SELENOMET-COOH PEPTIDE, ALPHA-HYDROXYFARNESYLPHOSPHONIC ACID, ... | | 著者 | Strickland, C.L, Windsor, W.T, Syto, R, Wang, L, Bond, R, Wu, Z, Schwartz, J, Le, H.V, Beese, L.S, Weber, P.C. | | 登録日 | 1999-04-27 | | 公開日 | 1999-06-18 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Crystal structure of farnesyl protein transferase complexed with a CaaX peptide and farnesyl diphosphate analogue
Biochemistry, 37, 1998
|
|
5HHH
 
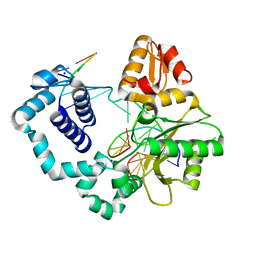 | | Structure of human DNA polymerase beta Host-Guest complexed with the control G for N7-CBZ-platination | | 分子名称: | DNA (5'-D(*CP*CP*GP*AP*CP*GP*GP*AP*GP*GP*AP*GP*CP*AP*GP*G)-3'), DNA (5'-D(P*CP*CP*TP*GP*CP*TP*CP*CP*TP*C)-3'), DNA (5'-D(P*GP*TP*CP*GP*G)-3'), ... | | 著者 | Koag, M.-C, Lee, S. | | 登録日 | 2016-01-11 | | 公開日 | 2017-01-18 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.363 Å) | | 主引用文献 | Synthesis, structure, and biological evaluation of a platinum-carbazole conjugate.
Chem Biol Drug Des, 91, 2018
|
|
7MMD
 
 | |
2XR5
 
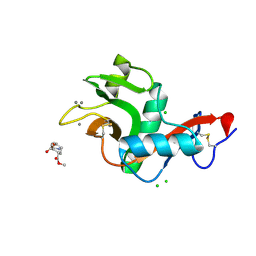 | | Crystal structure of the complex of the carbohydrate recognition domain of human DC-SIGN with pseudo dimannoside mimic. | | 分子名称: | CALCIUM ION, CD209 ANTIGEN, CHLORIDE ION, ... | | 著者 | Thepaut, M, Suitkeviciute, I, Sattin, S, Reina, J, Bernardi, A, Fieschi, F. | | 登録日 | 2010-09-10 | | 公開日 | 2011-10-19 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.42 Å) | | 主引用文献 | Structure of a Glycomimetic Ligand in the Carbohydrate Recognition Domain of C-Type Lectin Dc-Sign. Structural Requirements for Selectivity and Ligand Design.
J.Am.Chem.Soc., 135, 2013
|
|
3PLA
 
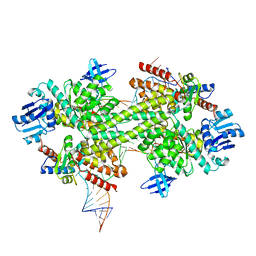 | | Crystal structure of a catalytically active substrate-bound box C/D RNP from Sulfolobus solfataricus | | 分子名称: | 50S ribosomal protein L7Ae, C/D guide RNA, Fibrillarin-like rRNA/tRNA 2'-O-methyltransferase, ... | | 著者 | Lin, J, Lai, S, Jia, R, Xu, A, Zhang, L, Lu, J, Ye, K. | | 登録日 | 2010-11-15 | | 公開日 | 2011-01-26 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (3.15 Å) | | 主引用文献 | Structural basis for site-specific ribose methylation by box C/D RNA protein complexes.
Nature, 469, 2011
|
|
2XX4
 
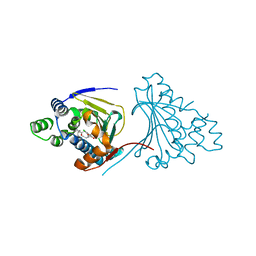 | | Macrolactone Inhibitor bound to HSP90 N-term | | 分子名称: | (E)-ETHYL 13-CHLORO-14,16-DIHYDROXY-1,11-DIOXO-1,2,3,4,7,8,9,10,11,12-DECAHYDROBENZO[C][1]AZACYCLOTETRADECINE-10-CARBOXYLATE, ATP-DEPENDENT MOLECULAR CHAPERONE HSP82 | | 著者 | Moody, C.J, Prodromou, C, Pearl, L.H, Roe, S.M. | | 登録日 | 2010-11-08 | | 公開日 | 2011-11-16 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.199 Å) | | 主引用文献 | Targeting the Hsp90 Molecular Chaperone with Novel Macrolactams. Synthesis, Structural, Binding, and Cellular Studies.
Acs Chem.Biol., 6, 2011
|
|
7WV3
 
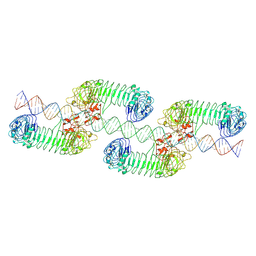 | | Toll-like receptor3 linear cluster | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, RNA (80-MER), ... | | 著者 | Lim, C.S, Jang, Y.H, Lee, G.Y, Han, G.M, Lee, J.O. | | 登録日 | 2022-02-09 | | 公開日 | 2022-11-16 | | 最終更新日 | 2022-12-07 | | 実験手法 | ELECTRON MICROSCOPY (2.26 Å) | | 主引用文献 | TLR3 forms a highly organized cluster when bound to a poly(I:C) RNA ligand.
Nat Commun, 13, 2022
|
|
1ZGI
 
 | | thrombin in complex with an oxazolopyridine inhibitor 21 | | 分子名称: | (R)-2-(2-(1H-1,2,4-TRIAZOL-1-YL)BENZYL)-N-(2,2-DIFLUORO-2-(PIPERIDIN-2-YL)ETHYL)OXAZOLO[4,5-C]PYRIDIN-4-AMINE, Hirudin, Thrombin | | 著者 | Deng, J.Z, McMasters, D.R, Rabbat, P.M, Williams, P.D, Coburn, C.A, Yan, Y, Kuo, L.C, Lewis, S.D, Lucas, B.J, Krueger, J.A, Strulovici, B, Vacca, J.P, Lyle, T.A, Burgey, C.S. | | 登録日 | 2005-04-21 | | 公開日 | 2005-09-27 | | 最終更新日 | 2013-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Development of an oxazolopyridine series of dual thrombin/factor Xa inhibitors via structure-guided lead optimization.
Bioorg.Med.Chem.Lett., 15, 2005
|
|
5DOQ
 
 | | The structure of bd oxidase from Geobacillus thermodenitrificans | | 分子名称: | Bd-type quinol oxidase subunit I, Bd-type quinol oxidase subunit II, CIS-HEME D HYDROXYCHLORIN GAMMA-SPIROLACTONE, ... | | 著者 | Safarian, S, Mueller, H, Rajendran, C, Preu, J, Ovchinnikov, S, Kusumoto, T, Hirose, T, Langer, J, Sakamoto, J, Michel, H. | | 登録日 | 2015-09-11 | | 公開日 | 2016-05-04 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (3.05 Å) | | 主引用文献 | Structure of a bd oxidase indicates similar mechanisms for membrane-integrated oxygen reductases.
Science, 352, 2016
|
|
