6ID0
 
 | | Cryo-EM structure of a human intron lariat spliceosome prior to Prp43 loaded (ILS1 complex) at 2.9 angstrom resolution | | 分子名称: | 116 kDa U5 small nuclear ribonucleoprotein component, CWF19-like protein 2, Cell division cycle 5-like protein, ... | | 著者 | Zhang, X, Zhan, X, Yan, C, Shi, Y. | | 登録日 | 2018-09-07 | | 公開日 | 2019-03-13 | | 最終更新日 | 2020-10-14 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Structures of the human spliceosomes before and after release of the ligated exon.
Cell Res., 29, 2019
|
|
6J6G
 
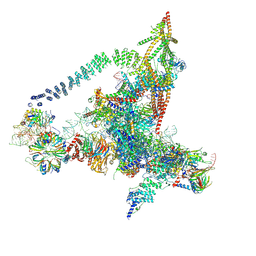 | | Cryo-EM structure of the yeast B*-a2 complex at an average resolution of 3.2 angstrom | | 分子名称: | ACT1 pre-mRNA, GUANOSINE-5'-TRIPHOSPHATE, INOSITOL HEXAKISPHOSPHATE, ... | | 著者 | Wan, R, Bai, R, Yan, C, Lei, J, Shi, Y. | | 登録日 | 2019-01-15 | | 公開日 | 2019-04-24 | | 最終更新日 | 2020-10-14 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Structures of the Catalytically Activated Yeast Spliceosome Reveal the Mechanism of Branching.
Cell, 177, 2019
|
|
6ID1
 
 | | Cryo-EM structure of a human intron lariat spliceosome after Prp43 loaded (ILS2 complex) at 2.9 angstrom resolution | | 分子名称: | 116 kDa U5 small nuclear ribonucleoprotein component, CWF19-like protein 2, Cell division cycle 5-like protein, ... | | 著者 | Zhang, X, Zhan, X, Yan, C, Shi, Y. | | 登録日 | 2018-09-07 | | 公開日 | 2019-03-13 | | 最終更新日 | 2024-10-09 | | 実験手法 | ELECTRON MICROSCOPY (2.86 Å) | | 主引用文献 | Structures of the human spliceosomes before and after release of the ligated exon.
Cell Res., 29, 2019
|
|
6J6H
 
 | | Cryo-EM structure of the yeast B*-a1 complex at an average resolution of 3.6 angstrom | | 分子名称: | ACT1 pre-mRNA, GUANOSINE-5'-TRIPHOSPHATE, INOSITOL HEXAKISPHOSPHATE, ... | | 著者 | Wan, R, Bai, R, Yan, C, Lei, J, Shi, Y. | | 登録日 | 2019-01-15 | | 公開日 | 2019-04-24 | | 最終更新日 | 2020-10-14 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Structures of the Catalytically Activated Yeast Spliceosome Reveal the Mechanism of Branching.
Cell, 177, 2019
|
|
6J6Q
 
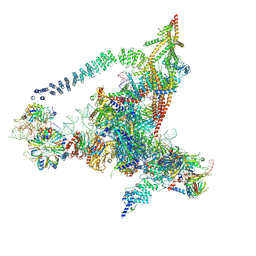 | | Cryo-EM structure of the yeast B*-b2 complex at an average resolution of 3.7 angstrom | | 分子名称: | GUANOSINE-5'-TRIPHOSPHATE, INOSITOL HEXAKISPHOSPHATE, MAGNESIUM ION, ... | | 著者 | Wan, R, Bai, R, Yan, C, Lei, J, Shi, Y. | | 登録日 | 2019-01-15 | | 公開日 | 2019-04-24 | | 最終更新日 | 2020-10-14 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Structures of the Catalytically Activated Yeast Spliceosome Reveal the Mechanism of Branching.
Cell, 177, 2019
|
|
6J6N
 
 | | Cryo-EM structure of the yeast B*-b1 complex at an average resolution of 3.86 angstrom | | 分子名称: | GUANOSINE-5'-TRIPHOSPHATE, INOSITOL HEXAKISPHOSPHATE, MAGNESIUM ION, ... | | 著者 | Wan, R, Bai, R, Yan, C, Lei, J, Shi, Y. | | 登録日 | 2019-01-15 | | 公開日 | 2019-04-24 | | 最終更新日 | 2020-10-14 | | 実験手法 | ELECTRON MICROSCOPY (3.86 Å) | | 主引用文献 | Structures of the Catalytically Activated Yeast Spliceosome Reveal the Mechanism of Branching.
Cell, 177, 2019
|
|
1BT1
 
 | | CATECHOL OXIDASE FROM IPOMOEA BATATAS (SWEET POTATOES) IN THE NATIVE CU(II)-CU(II) STATE | | 分子名称: | CU-O-CU LINKAGE, PROTEIN (CATECHOL OXIDASE) | | 著者 | Klabunde, T, Eicken, C, Sacchettini, J.C, Krebs, B. | | 登録日 | 1998-09-02 | | 公開日 | 1999-09-02 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Crystal structure of a plant catechol oxidase containing a dicopper center.
Nat.Struct.Biol., 5, 1998
|
|
1BT3
 
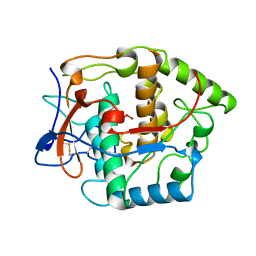 | | CATECHOL OXIDASE FROM IPOMOEA BATATAS (SWEET POTATOES) IN THE NATIVE CU(II)-CU(II) STATE | | 分子名称: | CU-O-CU LINKAGE, PROTEIN (CATECHOL OXIDASE) | | 著者 | Klabunde, T, Eicken, C, Sacchettini, J.C, Krebs, B. | | 登録日 | 1998-09-02 | | 公開日 | 1999-09-02 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Crystal structure of a plant catechol oxidase containing a dicopper center.
Nat.Struct.Biol., 5, 1998
|
|
1BUG
 
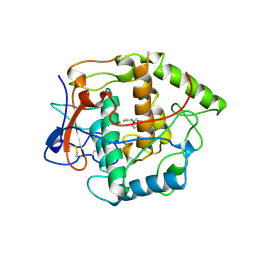 | | CATECHOL OXIDASE FROM IPOMOEA BATATAS (SWEET POTATOES)-INHIBITOR COMPLEX WITH PHENYLTHIOUREA (PTU) | | 分子名称: | COPPER (II) ION, N-PHENYLTHIOUREA, PROTEIN (CATECHOL OXIDASE) | | 著者 | Klabunde, T, Eicken, C, Sacchettini, J.C, Krebs, B. | | 登録日 | 1998-09-03 | | 公開日 | 1999-09-02 | | 最終更新日 | 2024-10-09 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Crystal structure of a plant catechol oxidase containing a dicopper center.
Nat.Struct.Biol., 5, 1998
|
|
1BT2
 
 | | CATECHOL OXIDASE FROM IPOMOEA BATATAS (SWEET POTATOES) IN THE REDUCED CU(I)-CU(I) STATE | | 分子名称: | CU-O-CU LINKAGE, PROTEIN (CATECHOL OXIDASE) | | 著者 | Klabunde, T, Eicken, C, Sacchettini, J.C, Krebs, B. | | 登録日 | 1998-09-02 | | 公開日 | 1999-09-02 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Crystal structure of a plant catechol oxidase containing a dicopper center.
Nat.Struct.Biol., 5, 1998
|
|
7TDZ
 
 | |
2WTM
 
 | | Est1E from Butyrivibrio proteoclasticus | | 分子名称: | EST1E, GLYCEROL, PHOSPHATE ION | | 著者 | Goldstone, D.C, Arcus, V.L. | | 登録日 | 2009-09-17 | | 公開日 | 2010-01-19 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Structural and Functional Characterization of a Promiscuous Feruloyl Esterase (Est1E) from the Rumen Bacterium Butyrivibrio Proteoclasticus.
Proteins, 78, 2010
|
|
4XMN
 
 | | Structure of the yeast coat nucleoporin complex, space group P212121 | | 分子名称: | Antibody 87 heavy chain, Antibody 87 light chain, Nucleoporin NUP120, ... | | 著者 | Stuwe, T, Correia, A.R, Lin, D.H, Paduch, M, Lu, V.T, Kossiakoff, A.A, Hoelz, A. | | 登録日 | 2015-01-14 | | 公開日 | 2015-03-25 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (7.6 Å) | | 主引用文献 | Nuclear pores. Architecture of the nuclear pore complex coat.
Science, 347, 2015
|
|
4XMM
 
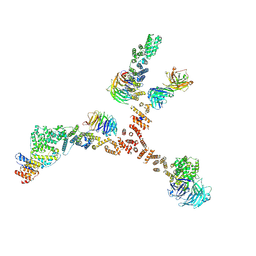 | | Structure of the yeast coat nucleoporin complex, space group C2 | | 分子名称: | Antibody 57 heavy chain, Antibody 57 light chain, Nucleoporin NUP120, ... | | 著者 | Stuwe, T, Correia, A.R, Lin, D.H, Paduch, M, Lu, V.T, Kossiakoff, A.A, Hoelz, A. | | 登録日 | 2015-01-14 | | 公開日 | 2015-03-25 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (7.384 Å) | | 主引用文献 | Nuclear pores. Architecture of the nuclear pore complex coat.
Science, 347, 2015
|
|
2N55
 
 | |
2WTN
 
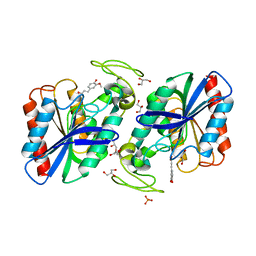 | |
5FKU
 
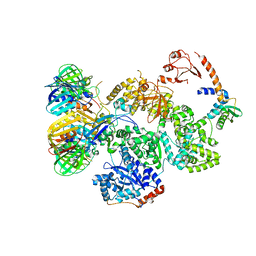 | | cryo-EM structure of the E. coli replicative DNA polymerase complex in DNA free state (DNA polymerase III alpha, beta, epsilon, tau complex) | | 分子名称: | DNA POLYMERASE III SUBUNIT ALPHA, DNA POLYMERASE III SUBUNIT BETA, DNA POLYMERASE III SUBUNIT EPSILON, ... | | 著者 | Fernandez-Leiro, R, Conrad, J, Scheres, S.H.W, Lamers, M.H. | | 登録日 | 2015-10-20 | | 公開日 | 2015-11-25 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (8.34 Å) | | 主引用文献 | cryo-EM structures of theE. colireplicative DNA polymerase reveal its dynamic interactions with the DNA sliding clamp, exonuclease andtau.
Elife, 4, 2015
|
|
2WB1
 
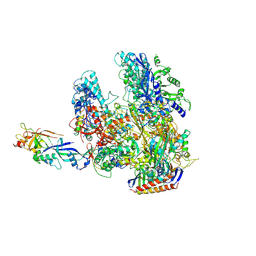 | | The complete structure of the archaeal 13-subunit DNA-directed RNA Polymerase | | 分子名称: | DNA-DIRECTED RNA POLYMERASE RPO10 SUBUNIT, DNA-DIRECTED RNA POLYMERASE RPO11 SUBUNIT, DNA-DIRECTED RNA POLYMERASE RPO12 SUBUNIT, ... | | 著者 | Korkhin, Y, Unligil, U.M, Littlefield, O, Nelson, P.J, Stuart, D.I, Sigler, P.B, Bell, S.D, Abrescia, N.G.A. | | 登録日 | 2009-02-19 | | 公開日 | 2009-05-19 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (3.52 Å) | | 主引用文献 | Evolution of Complex RNA Polymerase: The Complete Archaeal RNA Polymerase Structure
Plos Biol., 7, 2009
|
|
3B71
 
 | |
3BG1
 
 | |
4YCZ
 
 | |
3BG0
 
 | |
5GAN
 
 | | The overall structure of the yeast spliceosomal U4/U6.U5 tri-snRNP at 3.7 Angstrom | | 分子名称: | 13 kDa ribonucleoprotein-associated protein, GUANOSINE-5'-TRIPHOSPHATE, Pre-mRNA-processing factor 31, ... | | 著者 | Nguyen, T.H.D, Galej, W.P, Bai, X.C, Oubridge, C, Scheres, S.H.W, Newman, A.J, Nagai, K. | | 登録日 | 2015-12-15 | | 公開日 | 2016-01-27 | | 最終更新日 | 2024-05-15 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Cryo-EM structure of the yeast U4/U6.U5 tri-snRNP at 3.7 angstrom resolution.
Nature, 530, 2016
|
|
4Z0Z
 
 | |
6BOG
 
 | | Crystal structure of RapA, a Swi2/Snf2 protein that recycles RNA polymerase during transcription | | 分子名称: | RNA polymerase-associated protein RapA, SULFATE ION | | 著者 | Shaw, G.X, Gan, J, Zhou, Y.N, Zhang, R, Joachimiak, A, Jin, D.J, Ji, X. | | 登録日 | 2017-11-20 | | 公開日 | 2017-12-13 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (3.205 Å) | | 主引用文献 | Structure of RapA, a Swi2/Snf2 protein that recycles RNA polymerase during transcription.
Structure, 16, 2008
|
|
