5Y0U
 
 | |
2MIU
 
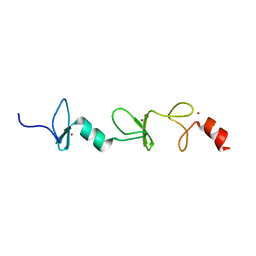 | | Structure of FHL2 LIM adaptor and its Interaction with Ski | | 分子名称: | Four and a half LIM domains protein 2, ZINC ION | | 著者 | Yang, Y, Sun, Y, Medrano, E.E, Tian, X, Weiss, M.A. | | 登録日 | 2013-12-20 | | 公開日 | 2014-01-15 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structure of FHL2 LIM adaptor and its Interaction with Ski
To be Published
|
|
8EQV
 
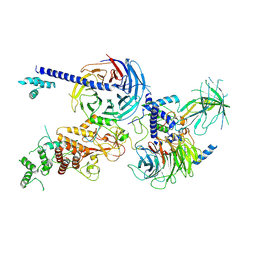 | | Cryo-EM structure of PRC2 in complex with the long isoform of AEBP2 | | 分子名称: | Histone-binding protein RBBP4, Histone-lysine N-methyltransferase EZH2, Polycomb protein EED, ... | | 著者 | Boudes, M, Zhang, Q, Flanigan, S.F, Davidovich, C. | | 登録日 | 2022-10-09 | | 公開日 | 2024-07-31 | | 実験手法 | ELECTRON MICROSCOPY (3.64 Å) | | 主引用文献 | To be updated
To Be Published
|
|
8FYH
 
 | | G4 RNA-mediated PRC2 dimer | | 分子名称: | G4 RNA, Histone-binding protein RBBP4, Histone-lysine N-methyltransferase EZH2, ... | | 著者 | Song, J, Kasinath, V. | | 登録日 | 2023-01-26 | | 公開日 | 2023-10-04 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Structural basis for inactivation of PRC2 by G-quadruplex RNA.
Science, 381, 2023
|
|
8TZX
 
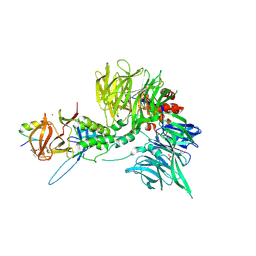 | | Ternary complex structure of Cereblon-DDB1 bound to WIZ(ZF7) and the molecular glue dWIZ-1 | | 分子名称: | (3S)-3-(5-{(1R)-1-[(2R)-1-ethylpiperidin-2-yl]ethoxy}-1-oxo-1,3-dihydro-2H-isoindol-2-yl)piperidine-2,6-dione, 1,2-ETHANEDIOL, DNA damage-binding protein 1, ... | | 著者 | Clifton, M.C, Ma, X, Ornelas, E. | | 登録日 | 2023-08-28 | | 公開日 | 2024-07-10 | | 最終更新日 | 2024-07-17 | | 実験手法 | X-RAY DIFFRACTION (3.15 Å) | | 主引用文献 | A molecular glue degrader of the WIZ transcription factor for fetal hemoglobin induction.
Science, 385, 2024
|
|
8W01
 
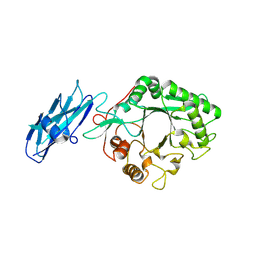 | |
8W04
 
 | |
8WRJ
 
 | | glycosyltransferase UGT74AN3 | | 分子名称: | 5-[(1R,2S,4R,6R,7R,10S,11S,14S,16R)-14-hydroxy-7,11-dimethyl-3-oxapentacyclo[8.8.0.02,4.02,7.011,16]octadecan-6-yl]pyran-2-one, Glycosyltransferase, URIDINE-5'-DIPHOSPHATE | | 著者 | Huang, W, Long, F. | | 登録日 | 2023-10-15 | | 公開日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Substrate Promiscuity, Crystal Structure, and Application of a Plant UDP-Glycosyltransferase UGT74AN3
Acs Catalysis, 14, 2024
|
|
8WRK
 
 | | glycosyltransferase UGT74AN3 | | 分子名称: | Glycosyltransferase, PICEATANNOL, URIDINE-5'-DIPHOSPHATE | | 著者 | Huang, W, Long, F. | | 登録日 | 2023-10-15 | | 公開日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.03 Å) | | 主引用文献 | Substrate Promiscuity, Crystal Structure, and Application of a Plant UDP-Glycosyltransferase UGT74AN3
Acs Catalysis, 14, 2024
|
|
8IND
 
 | | Crystal structure of UGT74AN3-UDP-RES | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5-[(1R,2S,4R,6R,7R,10S,11S,14S,16R)-14-hydroxy-7,11-dimethyl-3-oxapentacyclo[8.8.0.02,4.02,7.011,16]octadecan-6-yl]pyran-2-one, Glycosyltransferase, ... | | 著者 | Huang, W. | | 登録日 | 2023-03-09 | | 公開日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Substrate Promiscuity, Crystal Structure, and Application of a Plant UDP-Glycosyltransferase UGT74AN3
Acs Catalysis, 14, 2024
|
|
8INV
 
 | | Crystal structure of UGT74AN3-UDP-BUF | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Glycosyltransferase, URIDINE-5'-DIPHOSPHATE, ... | | 著者 | Long, F, Huang, W. | | 登録日 | 2023-03-10 | | 公開日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Substrate Promiscuity, Crystal Structure, and Application of a Plant UDP-Glycosyltransferase UGT74AN3
Acs Catalysis, 14, 2024
|
|
8INA
 
 | | Crystal structure of UGT74AN3-UDP | | 分子名称: | GLYCEROL, Glycosyltransferase, URIDINE-5'-DIPHOSPHATE | | 著者 | Long, F, Huang, W. | | 登録日 | 2023-03-09 | | 公開日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.86 Å) | | 主引用文献 | Substrate Promiscuity, Crystal Structure, and Application of a Plant UDP-Glycosyltransferase UGT74AN3
Acs Catalysis, 14, 2024
|
|
8INO
 
 | | Crystal structure of UGT74AN3 in complex UDP and PER | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-[(3S,5S,8S,9S,10R,13R,14S,17R)-10,13-dimethyl-3,5,14-tris(oxidanyl)-2,3,4,6,7,8,9,11,12,15,16,17-dodecahydro-1H-cyclopenta[a]phenanthren-17-yl]-2H-furan-5-one, Glycosyltransferase, ... | | 著者 | Long, F, Huang, W. | | 登録日 | 2023-03-10 | | 公開日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Substrate Promiscuity, Crystal Structure, and Application of a Plant UDP-Glycosyltransferase UGT74AN3
Acs Catalysis, 14, 2024
|
|
8I8Z
 
 | |
8INJ
 
 | | Crystal structure of UGT74AN3-UDP-DIG | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DIGITOXIGENIN, Glycosyltransferase, ... | | 著者 | Feng, L, Wei, H. | | 登録日 | 2023-03-10 | | 公開日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.76 Å) | | 主引用文献 | Crystal structure of UGT74AN3-UDP-DIG
To Be Published
|
|
8IN7
 
 | |
4A0F
 
 | | Structure of selenomethionine substituted bifunctional DAPA aminotransferase-dethiobiotin synthetase from Arabidopsis thaliana in its apo form. | | 分子名称: | ADENOSYLMETHIONINE-8-AMINO-7-OXONONANOATE AMINOTRANSFERASE, PYRIDOXAL-5'-PHOSPHATE, SULFATE ION | | 著者 | Cobessi, D, Dumas, R, Pautre, V, Meinguet, C, Ferrer, J.L, Alban, C. | | 登録日 | 2011-09-09 | | 公開日 | 2012-06-13 | | 実験手法 | X-RAY DIFFRACTION (2.714 Å) | | 主引用文献 | Biochemical and Structural Characterization of the Arabidopsis Bifunctional Enzyme Dethiobiotin Synthetase-Diaminopelargonic Acid Aminotransferase: Evidence for Substrate Channeling in Biotin Synthesis.
Plant Cell, 24, 2012
|
|
4A0R
 
 | | Structure of bifunctional DAPA aminotransferase-DTB synthetase from Arabidopsis thaliana bound to dethiobiotin (DTB). | | 分子名称: | 6-(5-METHYL-2-OXO-IMIDAZOLIDIN-4-YL)-HEXANOIC ACID, ADENOSYLMETHIONINE-8-AMINO-7-OXONONANOATE AMINOTRANSFERASE, L(+)-TARTARIC ACID, ... | | 著者 | Cobessi, D, Dumas, R, Pautre, V, Meinguet, C, Ferrer, J.L, Alban, C. | | 登録日 | 2011-09-12 | | 公開日 | 2012-06-13 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (2.68 Å) | | 主引用文献 | Biochemical and Structural Characterization of the Arabidopsis Bifunctional Enzyme Dethiobiotin Synthetase-Diaminopelargonic Acid Aminotransferase: Evidence for Substrate Channeling in Biotin Synthesis.
Plant Cell, 24, 2012
|
|
4A0H
 
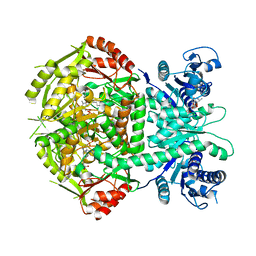 | | Structure of bifunctional DAPA aminotransferase-DTB synthetase from Arabidopsis thaliana bound to 7-keto 8-amino pelargonic acid (KAPA) | | 分子名称: | 7-KETO-8-AMINOPELARGONIC ACID, ADENOSYLMETHIONINE-8-AMINO-7-OXONONANOATE AMINOTRANSFERASE, L(+)-TARTARIC ACID, ... | | 著者 | Cobessi, D, Dumas, R, Pautre, V, Meinguet, C, Ferrer, J.L, Alban, C. | | 登録日 | 2011-09-09 | | 公開日 | 2012-06-13 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.808 Å) | | 主引用文献 | Biochemical and Structural Characterization of the Arabidopsis Bifunctional Enzyme Dethiobiotin Synthetase-Diaminopelargonic Acid Aminotransferase: Evidence for Substrate Channeling in Biotin Synthesis.
Plant Cell, 24, 2012
|
|
4A0G
 
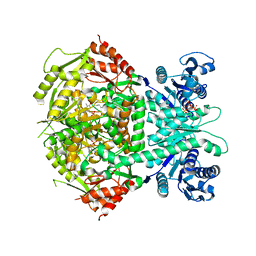 | | Structure of bifunctional DAPA aminotransferase-DTB synthetase from Arabidopsis thaliana in its apo form. | | 分子名称: | ADENOSYLMETHIONINE-8-AMINO-7-OXONONANOATE AMINOTRANSFERASE, MAGNESIUM ION, PYRIDOXAL-5'-PHOSPHATE, ... | | 著者 | Cobessi, D, Dumas, R, Pautre, V, Meinguet, C, Ferrer, J.L, Alban, C. | | 登録日 | 2011-09-09 | | 公開日 | 2012-06-13 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.502 Å) | | 主引用文献 | Biochemical and Structural Characterization of the Arabidopsis Bifunctional Enzyme Dethiobiotin Synthetase-Diaminopelargonic Acid Aminotransferase: Evidence for Substrate Channeling in Biotin Synthesis.
Plant Cell, 24, 2012
|
|
8P60
 
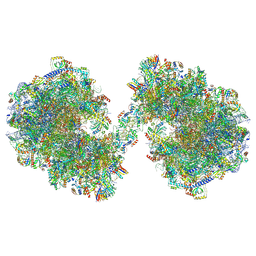 | | Spraguea lophii ribosome dimer | | 分子名称: | 40S Ribosomal protein S19, 40S ribosomal protein S0, 40S ribosomal protein S1, ... | | 著者 | Gil Diez, P, McLaren, M, Isupov, M.N, Daum, B, Conners, R, Williams, B. | | 登録日 | 2023-05-24 | | 公開日 | 2023-06-21 | | 最終更新日 | 2023-12-20 | | 実験手法 | ELECTRON MICROSCOPY (14.3 Å) | | 主引用文献 | CryoEM reveals that ribosomes in microsporidian spores are locked in a dimeric hibernating state.
Nat Microbiol, 8, 2023
|
|
8P5D
 
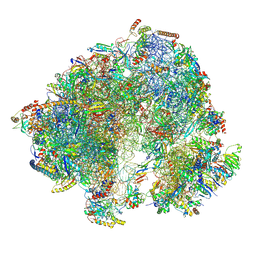 | | Spraguea lophii ribosome in the closed conformation by cryo sub tomogram averaging | | 分子名称: | 40S Ribosomal protein S19, 40S ribosomal protein S0, 40S ribosomal protein S10, ... | | 著者 | Gil Diez, P, McLaren, M, Isupov, M.N, Daum, B, Conners, R, Williams, B. | | 登録日 | 2023-05-23 | | 公開日 | 2023-06-21 | | 最終更新日 | 2023-12-20 | | 実験手法 | ELECTRON MICROSCOPY (10.8 Å) | | 主引用文献 | CryoEM reveals that ribosomes in microsporidian spores are locked in a dimeric hibernating state.
Nat Microbiol, 8, 2023
|
|
8I90
 
 | |
8I94
 
 | | Structure of flavone 4'-O-glucoside 7-O-glucosyltransferase from Nemophila menziesii, complex with luteolin | | 分子名称: | 2-(3,4-dihydroxyphenyl)-5,7-dihydroxy-4H-chromen-4-one, Glycosyltransferase, SULFATE ION | | 著者 | Murayama, K, Kato-Murayama, M, Shirouzu, M. | | 登録日 | 2023-02-06 | | 公開日 | 2024-02-14 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.43 Å) | | 主引用文献 | Molecular basis of ligand recognition specificity of flavone glucosyltransferases in Nemophila menziesii.
Arch.Biochem.Biophys., 753, 2024
|
|
7KSO
 
 | | Cryo-EM structure of PRC2:EZH1-AEBP2-JARID2 | | 分子名称: | Histone-binding protein RBBP4, Histone-lysine N-methyltransferase EZH1, Polycomb protein EED, ... | | 著者 | Grau, D.J, Armache, K.J. | | 登録日 | 2020-11-23 | | 公開日 | 2021-02-03 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Structures of monomeric and dimeric PRC2:EZH1 reveal flexible modules involved in chromatin compaction.
Nat Commun, 12, 2021
|
|
