7VAU
 
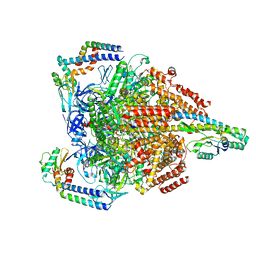 | | V1EG of V/A-ATPase from Thermus thermophilus at low ATP concentration, state2-2 | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | 著者 | Kishikawa, J, Nakanishi, A, Nakano, A, Saeki, S, Furuta, A, Kato, T, Mitsuoka, K, Yokoyama, K. | | 登録日 | 2021-08-30 | | 公開日 | 2022-07-13 | | 最終更新日 | 2024-06-19 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Structural snapshots of V/A-ATPase reveal the rotary catalytic mechanism of rotary ATPases.
Nat Commun, 13, 2022
|
|
7VAY
 
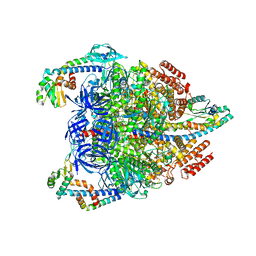 | | V1EG domain of V/A-ATPase from Thermus thermophilus at saturated ATP-gamma-S condition, state2 | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | 著者 | Kishikawa, J, Nakanishi, A, Nakano, A, Saeki, S, Furuta, A, Kato, T, Mitsuoka, K, Yokoyama, K. | | 登録日 | 2021-08-30 | | 公開日 | 2022-07-13 | | 最終更新日 | 2024-06-19 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Structural snapshots of V/A-ATPase reveal the rotary catalytic mechanism of rotary ATPases.
Nat Commun, 13, 2022
|
|
2PT1
 
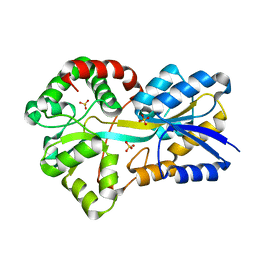 | | FutA1 Synechocystis PCC 6803 | | 分子名称: | Iron transport protein, SULFATE ION | | 著者 | Koropatkin, N.M, Randich, A.M, Bhattachryya-Pakrasi, M, Pakrasi, H.B, Smith, T.J. | | 登録日 | 2007-05-07 | | 公開日 | 2007-07-10 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | The structure of the iron-binding protein, FutA1, from Synechocystis 6803.
J.Biol.Chem., 282, 2007
|
|
1N0V
 
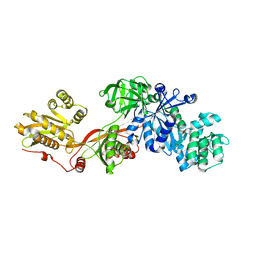 | | Crystal structure of elongation factor 2 | | 分子名称: | Elongation factor 2 | | 著者 | Joergensen, R, Ortiz, P.A, Carr-Schmid, A, Nissen, P, Kinzy, T.G, Andersen, G.R. | | 登録日 | 2002-10-15 | | 公開日 | 2002-11-27 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.85 Å) | | 主引用文献 | Two crystal structures demonstrate large conformational changes in the eukaryotic ribosomal translocase.
Nat.Struct.Biol., 10, 2003
|
|
3ZO3
 
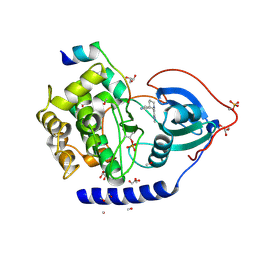 | | The Synthesis and Evaluation of Diazaspirocyclic Protein Kinase Inhibitors | | 分子名称: | 6-(2,9-DIAZASPIRO[5.5]UNDECAN-2-YL)-9H-PURINE, CAMP-DEPENDENT PROTEIN KINASE CATALYTIC SUBUNIT ALPHA, CAMP-DEPENDENT PROTEIN KINASE INHIBITOR ALPHA, ... | | 著者 | Allen, C.E, Chow, C.L, Caldwell, J.J, Westwood, I.M, van Montfort, R.L, Collins, I. | | 登録日 | 2013-02-20 | | 公開日 | 2013-03-06 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Synthesis and evaluation of heteroaryl substituted diazaspirocycles as scaffolds to probe the ATP-binding site of protein kinases.
Bioorg. Med. Chem., 21, 2013
|
|
6J5L
 
 | |
7Z4C
 
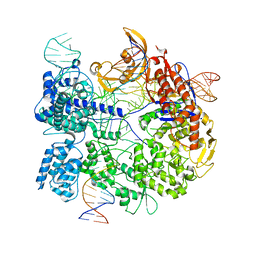 | | SpCas9 bound to 6 nucleotide complementary DNA substrate | | 分子名称: | CRISPR-associated endonuclease Cas9/Csn1, Non-target strand of 6 nucleotide complementary DNA substrate, Target strand of 6 nucleotide complementary DNA substrate, ... | | 著者 | Pacesa, M, Jinek, M. | | 登録日 | 2022-03-03 | | 公開日 | 2022-08-31 | | 最終更新日 | 2024-07-17 | | 実験手法 | ELECTRON MICROSCOPY (3.87 Å) | | 主引用文献 | R-loop formation and conformational activation mechanisms of Cas9.
Nature, 609, 2022
|
|
7Z4G
 
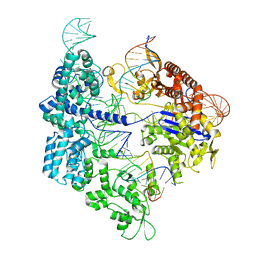 | | SpCas9 bound to 12-nucleotide complementary DNA substrate | | 分子名称: | CRISPR-associated endonuclease Cas9/Csn1, Non-target strand of 12-nucleotide complementary DNA substrate, Target strand of 12-nucleotide complementary DNA substrate, ... | | 著者 | Pacesa, M, Jinek, M. | | 登録日 | 2022-03-03 | | 公開日 | 2022-08-31 | | 最終更新日 | 2024-07-17 | | 実験手法 | ELECTRON MICROSCOPY (3.64 Å) | | 主引用文献 | R-loop formation and conformational activation mechanisms of Cas9.
Nature, 609, 2022
|
|
7Z4H
 
 | | SpCas9 bound to 14-nucleotide complementary DNA substrate | | 分子名称: | CRISPR-associated endonuclease Cas9/Csn1, Non-target strand of 14-nucleotide complementary DNA substrate, Target strand of 14-nucleotide complementary DNA substrate, ... | | 著者 | Pacesa, M, Jinek, M. | | 登録日 | 2022-03-03 | | 公開日 | 2022-08-31 | | 最終更新日 | 2024-07-17 | | 実験手法 | ELECTRON MICROSCOPY (3.49 Å) | | 主引用文献 | R-loop formation and conformational activation mechanisms of Cas9.
Nature, 609, 2022
|
|
7Z4K
 
 | | SpCas9 bound to 10-nucleotide complementary DNA substrate | | 分子名称: | CRISPR-associated endonuclease Cas9/Csn1, Non-target strand of 10-nucleotide complementary DNA substrate, Target strand of 10-nucleotide complementary DNA substrate, ... | | 著者 | Pacesa, M, Jinek, M. | | 登録日 | 2022-03-04 | | 公開日 | 2022-08-31 | | 最終更新日 | 2024-07-17 | | 実験手法 | ELECTRON MICROSCOPY (3.81 Å) | | 主引用文献 | R-loop formation and conformational activation mechanisms of Cas9.
Nature, 609, 2022
|
|
2BBD
 
 | | Crystal Structure of the STIV MCP | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, coat protein | | 著者 | Khayat, R. | | 登録日 | 2005-10-17 | | 公開日 | 2005-12-06 | | 最終更新日 | 2017-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.04 Å) | | 主引用文献 | Structure of an archaeal virus capsid protein reveals a common ancestry to eukaryotic and bacterial viruses.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
2VL0
 
 | |
1N0U
 
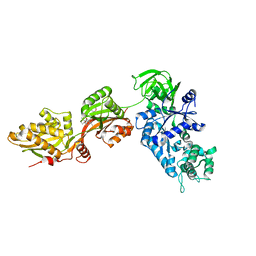 | | Crystal structure of yeast elongation factor 2 in complex with sordarin | | 分子名称: | Elongation factor 2, [1R-(1.ALPHA.,3A.BETA.,4.BETA.,4A.BETA.,7.BETA.,7A.ALPHA.,8A.BETA.)]8A-[(6-DEOXY-4-O-METHYL-BETA-D-ALTROPYRANOSYLOXY)METHYL]-4-FORMYL-4,4A,5,6,7,7A,8,8A-OCTAHYDRO-7-METHYL-3-(1-METHYLETHYL)-1,4-METHANO-S-INDACENE-3A(1H)-CARBOXYLIC ACID | | 著者 | Joergensen, R, Ortiz, P.A, Carr-Schmid, A, Nissen, P, Kinzy, T.G, Andersen, G.R. | | 登録日 | 2002-10-15 | | 公開日 | 2003-02-11 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.12 Å) | | 主引用文献 | Two crystal structures demonstrate large conformational changes in the eukaryotic ribosomal translocase.
Nat. Struct. Biol., 10, 2003
|
|
1GML
 
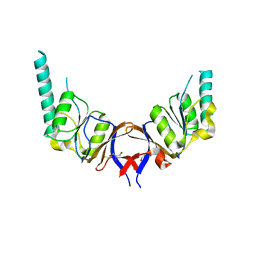 | | crystal structure of the mouse CCT gamma apical domain (triclinic) | | 分子名称: | GLYCEROL, T-COMPLEX PROTEIN 1 SUBUNIT GAMMA | | 著者 | Pappenberger, G, Wilsher, J.A, Roe, S.M, Willison, K.R, Pearl, L.H. | | 登録日 | 2001-09-17 | | 公開日 | 2002-06-18 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Crystal Structure of the Cct Gamma Apical Domain:: Implications for Substrate Binding to the Eukaryotic Cytosolic Chaperonin
J.Mol.Biol., 318, 2002
|
|
1GN1
 
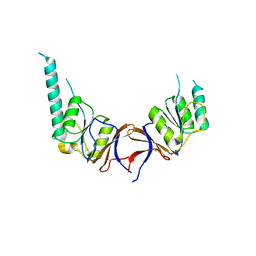 | | crystal structure of the mouse CCT gamma apical domain (monoclinic) | | 分子名称: | CALCIUM ION, CCT-GAMMA | | 著者 | Pappenberger, G, Wilsher, J.A, Roe, S.M, Willison, K.R, Pearl, L.H. | | 登録日 | 2001-10-01 | | 公開日 | 2002-06-18 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Crystal Structure of the Cct Gamma Apical Domain:: Implications for Substrate Binding to the Eukaryotic Cytosolic Chaperonin
J.Mol.Biol., 318, 2002
|
|
2VCO
 
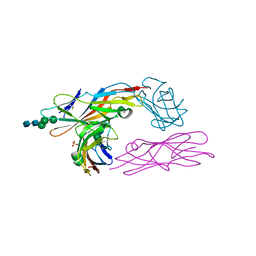 | | Crystal structure of the fimbrial adhesin FimH in complex with its high-mannose epitope | | 分子名称: | NICKEL (II) ION, PROTEIN FIMH, SULFATE ION, ... | | 著者 | Wellens, A, Garofalo, C, Nguyen, H, Wyns, L, De Greve, H, Hultgren, S.J, Bouckaert, J. | | 登録日 | 2007-09-26 | | 公開日 | 2008-05-13 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Intervening with Urinary Tract Infections Using Anti-Adhesives Based on the Crystal Structure of the Fimh-Oligomannose-3 Complex.
Plos One, 3, 2008
|
|
4MTA
 
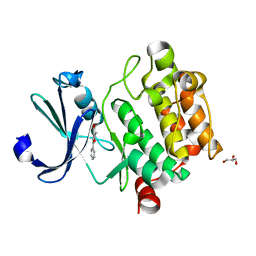 | |
2PT2
 
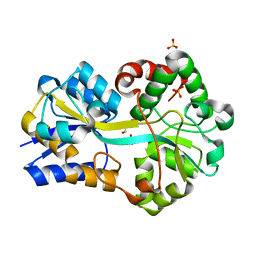 | | Structure of FutA1 with Iron(II) | | 分子名称: | 1,2-ETHANEDIOL, FE (II) ION, Iron transport protein, ... | | 著者 | Koropatkin, N.M, Randich, A.M, Bhattachryya-Pakrasi, M, Pakrasi, H.B, Smith, T.J. | | 登録日 | 2007-05-07 | | 公開日 | 2007-07-10 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | The structure of the iron-binding protein, FutA1, from Synechocystis 6803.
J.Biol.Chem., 282, 2007
|
|
5VWT
 
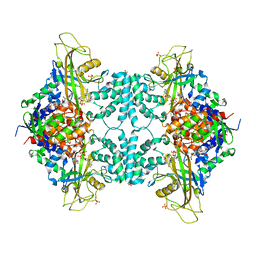 | |
2YKS
 
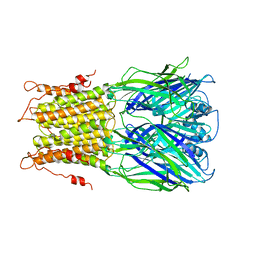 | |
4NT1
 
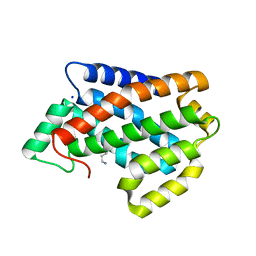 | | Crystal structure of apo-form of Arabidopsis ACD11 (accelerated-cell-death 11) at 1.8 Angstrom resolution | | 分子名称: | SODIUM ION, accelerated-cell-death 11 | | 著者 | Simanshu, D.K, Brown, R.E, Patel, D.J. | | 登録日 | 2013-11-29 | | 公開日 | 2014-02-05 | | 最終更新日 | 2014-02-19 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Arabidopsis Accelerated Cell Death 11, ACD11, Is a Ceramide-1-Phosphate Transfer Protein and Intermediary Regulator of Phytoceramide Levels.
Cell Rep, 6, 2014
|
|
4M3W
 
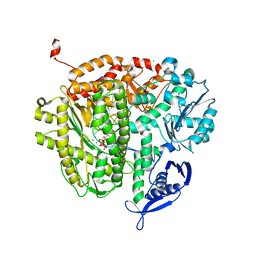 | |
2HW4
 
 | | Crystal structure of human phosphohistidine phosphatase | | 分子名称: | 14 kDa phosphohistidine phosphatase, FORMIC ACID | | 著者 | Busam, R.D, Thorsell, A.G, Arrowsmith, C, Berglund, H, Collins, R, Edwards, A, Ehn, M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Holmberg Schiavone, L, Hogbom, M, Kotenyova, T, Nilsson-Ehle, P, Nordlund, P, Nyman, T, Ogg, D, Stenmark, P, Sundstrom, M, Uppenberg, J, Van Den Berg, S, Weigelt, J, Persson, C, Hallberg, B.M, Structural Genomics Consortium (SGC) | | 登録日 | 2006-07-31 | | 公開日 | 2006-08-29 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | First structure of a eukaryotic phosphohistidine phosphatase
J.Biol.Chem., 281, 2006
|
|
6U5S
 
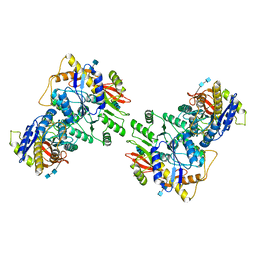 | |
6HMW
 
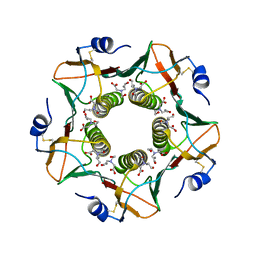 | |
