2F67
 
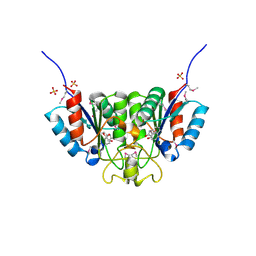 | | Crystal structure of Nucleoside 2-deoxyribosyltransferase from Trypanosoma brucei at 1.6 A resolution with BENZO[CD]INDOL-2(1H)-ONE bound | | 分子名称: | BENZO[CD]INDOL-2(1H)-ONE, GLYCEROL, Nucleoside 2-deoxyribosyltransferase, ... | | 著者 | Bosch, J, Robien, M.A, Hol, W.G.J, Structural Genomics of Pathogenic Protozoa Consortium (SGPP) | | 登録日 | 2005-11-28 | | 公開日 | 2005-12-06 | | 最終更新日 | 2024-10-09 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Using fragment cocktail crystallography to assist inhibitor design of Trypanosoma brucei nucleoside 2-deoxyribosyltransferase.
J.Med.Chem., 49, 2006
|
|
4D8Y
 
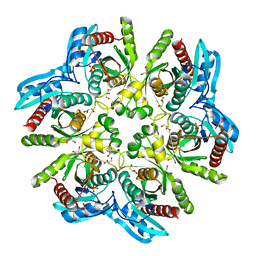 | | Crystal structure of the hexameric purine nucleoside phosphorylase from Bacillus subtilis in space group P212121 at pH 5.6 | | 分子名称: | GLYCEROL, Purine nucleoside phosphorylase deoD-type, SULFATE ION | | 著者 | Santos, C.R, Meza, A.N, Martins, N.H, Giuseppe, P.O, Murakami, M.T. | | 登録日 | 2012-01-11 | | 公開日 | 2012-09-26 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.61 Å) | | 主引用文献 | Insights into phosphate cooperativity and influence of substrate modifications on binding and catalysis of hexameric purine nucleoside phosphorylases.
Plos One, 7, 2012
|
|
4D8X
 
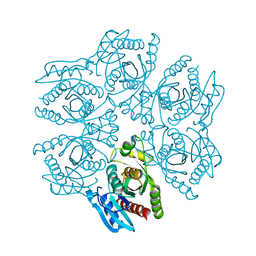 | | Crystal structure of the hexameric purine nucleoside phosphorylase from Bacillus subtilis in space group P6322 at pH 4.6 | | 分子名称: | Purine nucleoside phosphorylase deoD-type | | 著者 | Santos, C.R, Meza, A.N, Martins, N.H, Giuseppe, P.O, Murakami, M.T. | | 登録日 | 2012-01-11 | | 公開日 | 2012-09-26 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.65 Å) | | 主引用文献 | Insights into phosphate cooperativity and influence of substrate modifications on binding and catalysis of hexameric purine nucleoside phosphorylases.
Plos One, 7, 2012
|
|
4DA6
 
 | | Crystal structure of the hexameric purine nucleoside phosphorylase from Bacillus subtilis in complex with ganciclovir | | 分子名称: | 9-(1,3-DIHYDROXY-PROPOXYMETHANE)GUANINE, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | 著者 | Martins, N.H, Giuseppe, P.O, Meza, A.N, Murakami, M.T. | | 登録日 | 2012-01-12 | | 公開日 | 2012-09-26 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Insights into phosphate cooperativity and influence of substrate modifications on binding and catalysis of hexameric purine nucleoside phosphorylases.
Plos One, 7, 2012
|
|
4D8V
 
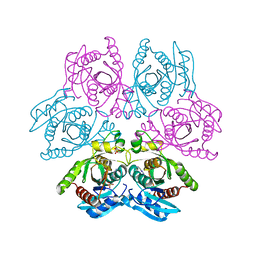 | | Crystal structure of the hexameric purine nucleoside phosphorylase from Bacillus subtilis at pH 4.2 | | 分子名称: | ADENINE, Purine nucleoside phosphorylase deoD-type, SULFATE ION | | 著者 | Santos, C.R, Meza, A.N, Martins, N.H, Giuseppe, P.O, Murakami, M.T. | | 登録日 | 2012-01-11 | | 公開日 | 2012-09-26 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | Insights into phosphate cooperativity and influence of substrate modifications on binding and catalysis of hexameric purine nucleoside phosphorylases.
Plos One, 7, 2012
|
|
4D98
 
 | | Crystal structure of the hexameric purine nucleoside phosphorylase from Bacillus subtilis in space group H32 at pH 7.5 | | 分子名称: | CHLORIDE ION, GLYCEROL, Purine nucleoside phosphorylase deoD-type, ... | | 著者 | Santos, C.R, Meza, A.N, Martins, N.H, Giuseppe, P.O, Murakami, M.T. | | 登録日 | 2012-01-11 | | 公開日 | 2012-09-26 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Insights into phosphate cooperativity and influence of substrate modifications on binding and catalysis of hexameric purine nucleoside phosphorylases.
Plos One, 7, 2012
|
|
4D9H
 
 | | Crystal structure of the hexameric purine nucleoside phosphorylase from Bacillus subtilis in complex with adenosine | | 分子名称: | ADENOSINE, Purine nucleoside phosphorylase deoD-type | | 著者 | Giuseppe, P.O, Martins, N.H, Meza, A.N, Murakami, M.T. | | 登録日 | 2012-01-11 | | 公開日 | 2012-09-26 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.91 Å) | | 主引用文献 | Insights into phosphate cooperativity and influence of substrate modifications on binding and catalysis of hexameric purine nucleoside phosphorylases.
Plos One, 7, 2012
|
|
4DA0
 
 | | Crystal structure of the hexameric purine nucleoside phosphorylase from Bacillus subtilis in complex with 2'-deoxyguanosine | | 分子名称: | 2'-DEOXY-GUANOSINE, CHLORIDE ION, Purine nucleoside phosphorylase deoD-type | | 著者 | Martins, N.H, Giuseppe, P.O, Meza, A.N, Murakami, M.T. | | 登録日 | 2012-01-12 | | 公開日 | 2012-09-26 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.95 Å) | | 主引用文献 | Insights into phosphate cooperativity and influence of substrate modifications on binding and catalysis of hexameric purine nucleoside phosphorylases.
Plos One, 7, 2012
|
|
5DK6
 
 | | CRYSTAL STRUCTURE OF A 5'-METHYLTHIOADENOSINE/S-ADENOSYLHOMOCYSTEINE (MTA/SAH) NUCLEOSIDASE (MTAN) FROM COLWELLIA PSYCHRERYTHRAEA 34H (CPS_4743, TARGET PSI-029300) IN COMPLEX WITH ADENINE AT 2.27 A RESOLUTION | | 分子名称: | 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase, ADENINE, GLYCINE | | 著者 | Himmel, D.M, Bhosle, R, Toro, R, Ahmed, M, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Love, J, Fiser, A, Seidel, R.D, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | 登録日 | 2015-09-03 | | 公開日 | 2015-11-04 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (2.27 Å) | | 主引用文献 | CRYSTAL STRUCTURE OF A 5'-METHYLTHIOADENOSINE/S-ADENOSYLHOMOCYSTEINE (MTA/SAH)NUCLEOSIDASE (MTAN) FROM COLWELLIA PSYCHRERYTHRAEA 34H (CPS_4743, TARGET PSI-029300) IN COMPLEX WITH ADENINE AT 2.27 A RESOLUTION
To be published
|
|
6GFM
 
 | | Crystal structure of the Escherichia coli nucleosidase PpnN (pppGpp-form) | | 分子名称: | Pyrimidine/purine nucleotide 5'-monophosphate nucleosidase, guanosine 5'-(tetrahydrogen triphosphate) 3'-(trihydrogen diphosphate) | | 著者 | Zhang, Y, Baerentsen, R.L, Gerdes, K, Brodersen, D.E. | | 登録日 | 2018-05-01 | | 公開日 | 2019-04-24 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.77 Å) | | 主引用文献 | (p)ppGpp Regulates a Bacterial Nucleosidase by an Allosteric Two-Domain Switch.
Mol.Cell, 74, 2019
|
|
6GFL
 
 | |
2F2T
 
 | | Crystal structure of Nucleoside 2-deoxyribosyltransferase from Trypanosoma brucei at 1.7 A resolution with 5-Aminoisoquinoline bound | | 分子名称: | GLYCEROL, ISOQUINOLIN-5-AMINE, Nucleoside 2-deoxyribosyltransferase, ... | | 著者 | Bosch, J, Robien, M.A, Hol, W.G.J, Structural Genomics of Pathogenic Protozoa Consortium (SGPP) | | 登録日 | 2005-11-17 | | 公開日 | 2005-11-22 | | 最終更新日 | 2024-11-13 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Using fragment cocktail crystallography to assist inhibitor design of Trypanosoma brucei nucleoside 2-deoxyribosyltransferase.
J.Med.Chem., 49, 2006
|
|
6F4X
 
 | |
1Z34
 
 | | Crystal structure of Trichomonas vaginalis purine nucleoside phosphorylase complexed with 2-fluoro-2'-deoxyadenosine | | 分子名称: | 5-(6-AMINO-2-FLUORO-PURIN-9-YL)-2-HYDROXYMETHYL-TETRAHYDRO-FURAN-3-OL, purine nucleoside phosphorylase | | 著者 | Zhang, Y, Wang, W.H, Wu, S.W, Wang, C.C, Ealick, S.E. | | 登録日 | 2005-03-10 | | 公開日 | 2005-03-29 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Identification of a subversive substrate of Trichomonas vaginalis purine nucleoside phosphorylase and the crystal structure of the enzyme-substrate complex.
J.Biol.Chem., 280, 2005
|
|
1Z36
 
 | | Crystal structure of Trichomonas vaginalis purine nucleoside phosphorylase complexed with formycin A | | 分子名称: | (1S)-1-(7-amino-1H-pyrazolo[4,3-d]pyrimidin-3-yl)-1,4-anhydro-D-ribitol, purine nucleoside phosphorylase | | 著者 | Zhang, Y, Wang, W.H, Wu, S.W, Wang, C.C, Ealick, S.E. | | 登録日 | 2005-03-10 | | 公開日 | 2005-03-29 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Identification of a subversive substrate of Trichomonas vaginalis purine nucleoside phosphorylase and the crystal structure of the enzyme-substrate complex.
J.Biol.Chem., 280, 2005
|
|
1Z33
 
 | | Crystal structure of Trichomonas vaginalis purine nucleoside phosphorylase | | 分子名称: | purine nucleoside phosphorylase | | 著者 | Zhang, Y, Wang, W.H, Wu, S.W, Wang, C.C, Ealick, S.E. | | 登録日 | 2005-03-10 | | 公開日 | 2005-03-29 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Identification of a subversive substrate of Trichomonas vaginalis purine nucleoside phosphorylase and the crystal structure of the enzyme-substrate complex.
J.Biol.Chem., 280, 2005
|
|
1Z39
 
 | | Crystal structure of Trichomonas vaginalis purine nucleoside phosphorylase complexed with 2'-deoxyinosine | | 分子名称: | 2'-DEOXYINOSINE, purine nucleoside phosphorylase | | 著者 | Zhang, Y, Wang, W.H, Wu, S.W, Wang, C.C, Ealick, S.E. | | 登録日 | 2005-03-10 | | 公開日 | 2005-03-29 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Identification of a subversive substrate of Trichomonas vaginalis purine nucleoside phosphorylase and the crystal structure of the enzyme-substrate complex.
J.Biol.Chem., 280, 2005
|
|
1Z38
 
 | | Crystal structure of Trichomonas vaginalis purine nucleoside phosphorylase complexed with inosine | | 分子名称: | INOSINE, purine nucleoside phosphorylase | | 著者 | Zhang, Y, Wang, W.H, Wu, S.W, Wang, C.C, Ealick, S.E. | | 登録日 | 2005-03-10 | | 公開日 | 2005-03-29 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Identification of a subversive substrate of Trichomonas vaginalis purine nucleoside phosphorylase and the crystal structure of the enzyme-substrate complex.
J.Biol.Chem., 280, 2005
|
|
2JJ8
 
 | |
1Z37
 
 | | Crystal structure of Trichomonas vaginalis purine nucleoside phosphorylase complexed with adenosine | | 分子名称: | ADENOSINE, purine nucleoside phosphorylase | | 著者 | Zhang, Y, Wang, W.H, Wu, S.W, Wang, C.C, Ealick, S.E. | | 登録日 | 2005-03-10 | | 公開日 | 2005-03-29 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Identification of a subversive substrate of Trichomonas vaginalis purine nucleoside phosphorylase and the crystal structure of the enzyme-substrate complex.
J.Biol.Chem., 280, 2005
|
|
1Z35
 
 | | Crystal structure of Trichomonas vaginalis purine nucleoside phosphorylase complexed with 2-fluoroadenosine | | 分子名称: | 2-(6-AMINO-2-FLUORO-PURIN-9-YL)-5-HYDROXYMETHYL-TETRAHYDRO-FURAN-3,4-DIOL, purine nucleoside phosphorylase | | 著者 | Zhang, Y, Wang, W.H, Wu, S.W, Wang, C.C, Ealick, S.E. | | 登録日 | 2005-03-10 | | 公開日 | 2005-03-29 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Identification of a subversive substrate of Trichomonas vaginalis purine nucleoside phosphorylase and the crystal structure of the enzyme-substrate complex.
J.Biol.Chem., 280, 2005
|
|
6GP1
 
 | | Structure of mEos4b in the red long-lived dark state | | 分子名称: | Green to red photoconvertible GFP-like protein EosFP | | 著者 | De Zitter, E, Adam, V, Byrdin, M, Van Meervelt, L, Dedecker, P, Bourgeois, D. | | 登録日 | 2018-06-04 | | 公開日 | 2019-05-22 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.504 Å) | | 主引用文献 | Mechanistic investigation of mEos4b reveals a strategy to reduce track interruptions in sptPALM.
Nat.Methods, 16, 2019
|
|
6GOZ
 
 | | Structure of mEos4b in the green long-lived dark state | | 分子名称: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | 著者 | De Zitter, E, Adam, V, Byrdin, M, Van Meervelt, L, Dedecker, P, Bourgeois, D. | | 登録日 | 2018-06-04 | | 公開日 | 2019-11-13 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (2.406 Å) | | 主引用文献 | Mechanistic Investigations of Green mEos4b Reveal a Dynamic Long-Lived Dark State.
J.Am.Chem.Soc., 2020
|
|
4PR3
 
 | | Crystal structure of Brucella melitensis 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase | | 分子名称: | 5'-methylthioadenosine nucleosidase / s-adenosylhomocysteine nucleosidase, ADENINE, GLYCEROL, ... | | 著者 | Zhang, X.C, Kang, X.S, Zhao, Y, Jiang, D.H, Li, X.M, Chen, Z.L. | | 登録日 | 2014-03-05 | | 公開日 | 2014-04-30 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (2.606 Å) | | 主引用文献 | Crystal structure and biochemical studies of Brucella melitensis 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase
Biochem.Biophys.Res.Commun., 446, 2014
|
|
2A0K
 
 | | Crystal structure of Nucleoside 2-deoxyribosyltransferase from Trypanosoma brucei at 1.8 A resolution | | 分子名称: | GLYCEROL, Nucleoside 2-deoxyribosyltransferase, SULFATE ION | | 著者 | Bosch, J, Robien, M.A, Hol, W.G.J, Structural Genomics of Pathogenic Protozoa Consortium (SGPP) | | 登録日 | 2005-06-16 | | 公開日 | 2005-07-26 | | 最終更新日 | 2024-10-09 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Using fragment cocktail crystallography to assist inhibitor design of Trypanosoma brucei nucleoside 2-deoxyribosyltransferase.
J.Med.Chem., 49, 2006
|
|
