8F5K
 
 | |
8F5L
 
 | | Azurin from Pseudomonas aeruginosa, Y72F/Y108F/F110L mutant | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Azurin, COPPER (II) ION | | 著者 | Zeug, M, Offenbacher, A.R, Choe, J. | | 登録日 | 2022-11-14 | | 公開日 | 2023-01-04 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.15 Å) | | 主引用文献 | Electrochemical and Structural Study of the Buried Tryptophan in Azurin: Effects of Hydration and Polarity on the Redox Potential of W48.
J.Phys.Chem.B, 127, 2023
|
|
3AA1
 
 | | Crystal structure of Actin capping protein in complex with the Cp-binding motif derived from CKIP-1 | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 23mer peptide from Pleckstrin homology domain-containing family O member 1, F-actin-capping protein subunit alpha-1, ... | | 著者 | Takeda, S, Minakata, S, Narita, A, Kitazawa, M, Yamakuni, T, Maeda, Y, Nitanai, Y. | | 登録日 | 2009-11-11 | | 公開日 | 2010-08-04 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Two distinct mechanisms for actin capping protein regulation--steric and allosteric inhibition
Plos Biol., 8, 2010
|
|
8IYQ
 
 | | Structure of CbCas9 bound to 20-nucleotide complementary DNA substrate | | 分子名称: | NTS, TS, deadCbCas9, ... | | 著者 | Zhang, S, Lin, S, Liu, J.J.G. | | 登録日 | 2023-04-05 | | 公開日 | 2024-06-05 | | 最終更新日 | 2024-06-26 | | 実験手法 | ELECTRON MICROSCOPY (2.46 Å) | | 主引用文献 | Pro-CRISPR PcrIIC1-associated Cas9 system for enhanced bacterial immunity.
Nature, 630, 2024
|
|
8K0S
 
 | |
8K15
 
 | |
8K0R
 
 | |
8K0Q
 
 | |
8K0P
 
 | |
1GSS
 
 | | THREE-DIMENSIONAL STRUCTURE OF CLASS PI GLUTATHIONE S-TRANSFERASE FROM HUMAN PLACENTA IN COMPLEX WITH S-HEXYLGLUTATHIONE AT 2.8 ANGSTROMS RESOLUTION | | 分子名称: | GLUTATHIONE S-TRANSFERASE, L-gamma-glutamyl-S-hexyl-L-cysteinylglycine | | 著者 | Reinemer, P, Dirr, H.W, Ladenstein, R, Lobello, M, Federici, G, Huber, R, Parker, M.W. | | 登録日 | 1992-05-28 | | 公開日 | 1994-01-31 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Three-dimensional structure of class pi glutathione S-transferase from human placenta in complex with S-hexylglutathione at 2.8 A resolution.
J.Mol.Biol., 227, 1992
|
|
7VP9
 
 | | Crystal structure of human ClpP in complex with ZG111 | | 分子名称: | (6S,9aS)-N-[(4-bromophenyl)methyl]-6-[(2S)-butan-2-yl]-8-(naphthalen-1-ylmethyl)-4,7-bis(oxidanylidene)-3,6,9,9a-tetrahydro-2H-pyrazino[1,2-a]pyrimidine-1-carboxamide, ATP-dependent Clp protease proteolytic subunit, mitochondrial, ... | | 著者 | Wang, P.Y, Gan, J.H, Yang, C.-G. | | 登録日 | 2021-10-15 | | 公開日 | 2022-06-08 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.552 Å) | | 主引用文献 | Aberrant human ClpP activation disturbs mitochondrial proteome homeostasis to suppress pancreatic ductal adenocarcinoma.
Cell Chem Biol, 29, 2022
|
|
7VPU
 
 | |
7VPS
 
 | |
7VPR
 
 | |
7VPT
 
 | |
7WKJ
 
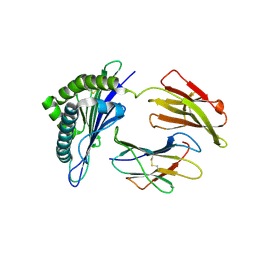 | | A COVID-19 T-cell response detection method based on a newly identified human CD8+ T cell epitope from SARS-CoV-2-Hubei Province, 2021. | | 分子名称: | Beta-2-microglobulin, LYS-THR-PHE-PRO-PRO-THR-GLU-PRO-LYS, MHC class I antigen | | 著者 | Zhang, J, Lu, D, Li, M, Liu, M.S, Yao, S.J, Zhan, J.B, Liu, J, Gao, G.F. | | 登録日 | 2022-01-10 | | 公開日 | 2022-03-02 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | A COVID-19 T-Cell Response Detection Method Based on a Newly Identified Human CD8 + T Cell Epitope from SARS-CoV-2 - Hubei Province, China, 2021.
China CDC Wkly, 4, 2022
|
|
1GTI
 
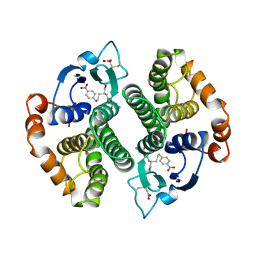 | | MODIFIED GLUTATHIONE S-TRANSFERASE (PI) COMPLEXED WITH S (P-NITROBENZYL)GLUTATHIONE | | 分子名称: | GLUTATHIONE S-TRANSFERASE, S-(P-NITROBENZYL)GLUTATHIONE | | 著者 | Vega, M.C, Coll, M. | | 登録日 | 1998-01-09 | | 公開日 | 1999-03-02 | | 最終更新日 | 2023-08-09 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | The three-dimensional structure of Cys-47-modified mouse liver glutathione S-transferase P1-1. Carboxymethylation dramatically decreases the affinity for glutathione and is associated with a loss of electron density in the alphaB-310B region.
J.Biol.Chem., 273, 1998
|
|
1GSY
 
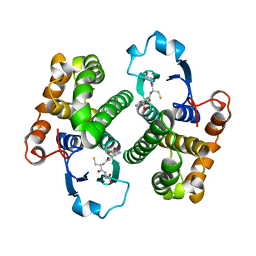 | | GLUTATHIONE S-TRANSFERASE YFYF, CLASS PI, COMPLEXED WITH GLUTATHIONE | | 分子名称: | GLUTATHIONE, GLUTATHIONE S-TRANSFERASE CLASS PI | | 著者 | Parraga, A, Garcia-Saez, I, Coll, M. | | 登録日 | 1996-10-25 | | 公開日 | 1997-11-19 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (2.44 Å) | | 主引用文献 | The three-dimensional structure of a class-Pi glutathione S-transferase complexed with glutathione: the active-site hydration provides insights into the reaction mechanism.
Biochem.J., 333 ( Pt 3), 1998
|
|
1GLQ
 
 | |
7WBG
 
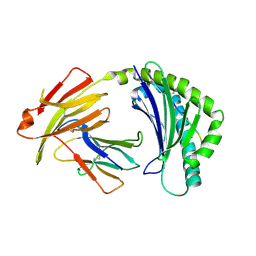 | | BF2*1901/RY8 | | 分子名称: | ARG-ARG-ARG-GLU-GLN-THR-ASP-TYR, Beta-2-microglobulin, MHC class I alpha chain 2 | | 著者 | Liu, W.J. | | 登録日 | 2021-12-16 | | 公開日 | 2022-12-21 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | A Wider and Deeper Peptide-Binding Groove for the Class I Molecules from B15 Compared with B19 Chickens Correlates with Relative Resistance to Marek's Disease.
J Immunol., 210, 2023
|
|
7X99
 
 | |
7XJT
 
 | |
7XB5
 
 | |
7WWG
 
 | | Crystal structure of Saccharomyces cerevisiae Sfh2 complexed with phosphatidylinositol in an open conformation | | 分子名称: | (1R)-2-{[(S)-hydroxy{[(1S,2R,3R,4S,5S,6R)-2,3,4,5,6-pentahydroxycyclohexyl]oxy}phosphoryl]oxy}-1-[(octadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, Phosphatidylinositol transfer protein CSR1 | | 著者 | Chen, L, Tan, L, Im, Y.J. | | 登録日 | 2022-02-12 | | 公開日 | 2022-07-13 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (3.4 Å) | | 主引用文献 | Structural basis of ligand recognition and transport by Sfh2, a yeast phosphatidylinositol transfer protein of the Sec14 superfamily.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7WWE
 
 | |
