6PFC
 
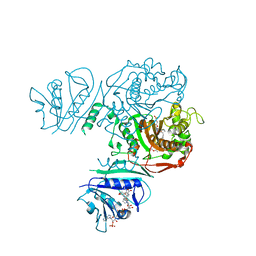 | | Crystal structure of TS-DHFR from Cryptosporidium hominis in complex with NADPH, FdUMP and 2-(2-(4-((2-amino-4-oxo-4,7-dihydro-3H-pyrrolo[2,3-d]pyrimidin-5-yl)methyl)benzamido)-4-methoxyphenyl)acetic acid | | 分子名称: | 5-FLUORO-2'-DEOXYURIDINE-5'-MONOPHOSPHATE, Bifunctional dihydrofolate reductase-thymidylate synthase, METHOTREXATE, ... | | 著者 | Czyzyk, D.J, Valhondo, M, Jorgensen, W.L, Anderson, K.S. | | 登録日 | 2019-06-21 | | 公開日 | 2019-10-16 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.689 Å) | | 主引用文献 | Structure activity relationship towards design of cryptosporidium specific thymidylate synthase inhibitors.
Eur.J.Med.Chem., 183, 2019
|
|
6PF5
 
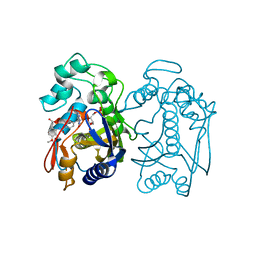 | | Crystal structure of human thymidylate synthase Delta (7-29) in complex with dUMP and 2-(2-(4-((2-amino-4-oxo-4,7-dihydro-3H-pyrrolo[2,3-d]pyrimidin-5-yl)methyl)benzamido)-4-methoxyphenyl)acetic acid | | 分子名称: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, Thymidylate synthase, [2-({4-[(2-amino-4-oxo-4,7-dihydro-3H-pyrrolo[2,3-d]pyrimidin-5-yl)methyl]benzene-1-carbonyl}amino)-4-methoxyphenyl]acetic acid | | 著者 | Czyzyk, D.J, Valhondo, M, Jorgensen, W.L, Anderson, K.S. | | 登録日 | 2019-06-21 | | 公開日 | 2019-10-02 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.39 Å) | | 主引用文献 | Structure activity relationship towards design of cryptosporidium specific thymidylate synthase inhibitors.
Eur.J.Med.Chem., 183, 2019
|
|
6PFF
 
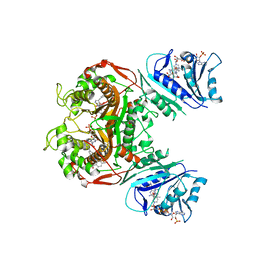 | | Crystal structure of TS-DHFR from Cryptosporidium hominis in complex with NADPH, FdUMP and 2-(4-((2-amino-4-oxo-4,7-dihydro-3H-pyrrolo[2,3-d]pyrimidin-5-yl)methyl)benzamido)-4-cyanobenzoic acid. | | 分子名称: | 2-({4-[(2-amino-4-oxo-4,7-dihydro-3H-pyrrolo[2,3-d]pyrimidin-5-yl)methyl]benzene-1-carbonyl}amino)-4-cyanobenzoic acid, 5-FLUORO-2'-DEOXYURIDINE-5'-MONOPHOSPHATE, Bifunctional dihydrofolate reductase-thymidylate synthase, ... | | 著者 | Czyzyk, D.J, Valhondo, M, Jorgensen, W.L, Anderson, K.S. | | 登録日 | 2019-06-21 | | 公開日 | 2019-10-02 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.982 Å) | | 主引用文献 | Structure activity relationship towards design of cryptosporidium specific thymidylate synthase inhibitors.
Eur.J.Med.Chem., 183, 2019
|
|
6Q0W
 
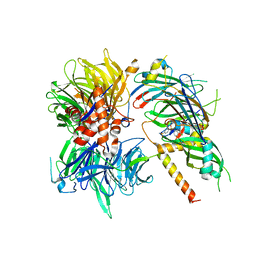 | | Structure of DDB1-DDA1-DCAF15 complex bound to Indisulam and RBM39 | | 分子名称: | DDB1- and CUL4-associated factor 15, DET1- and DDB1-associated protein 1, DNA damage-binding protein 1, ... | | 著者 | Faust, T, Yoon, H, Nowak, R.P, Donovan, K.A, Li, Z, Cai, Q, Eleuteri, N.A, Zhang, T, Gray, N.S, Fischer, E.S. | | 登録日 | 2019-08-02 | | 公開日 | 2019-11-13 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Structural complementarity facilitates E7820-mediated degradation of RBM39 by DCAF15.
Nat.Chem.Biol., 16, 2020
|
|
6Q0R
 
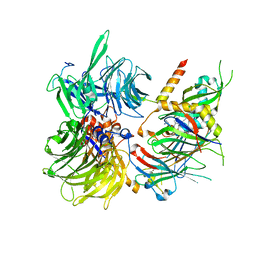 | | Structure of DDB1-DDA1-DCAF15 complex bound to E7820 and RBM39 | | 分子名称: | 3-cyano-N-(3-cyano-4-methyl-1H-indol-7-yl)benzene-1-sulfonamide, DDB1- and CUL4-associated factor 15, DET1- and DDB1-associated protein 1, ... | | 著者 | Faust, T, Yoon, H, Nowak, R.P, Donovan, K.A, Li, Z, Cai, Q, Eleuteri, N.A, Zhang, T, Gray, N.S, Fischer, E.S. | | 登録日 | 2019-08-02 | | 公開日 | 2019-11-13 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Structural complementarity facilitates E7820-mediated degradation of RBM39 by DCAF15.
Nat.Chem.Biol., 16, 2020
|
|
6Q4B
 
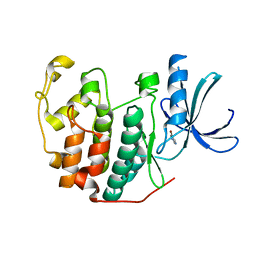 | | CDK2 in complex with FragLite13 | | 分子名称: | 5-bromanylpyrimidine, Cyclin-dependent kinase 2 | | 著者 | Wood, D.J, Martin, M.P, Noble, M.E.M. | | 登録日 | 2018-12-05 | | 公開日 | 2019-03-20 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.12 Å) | | 主引用文献 | FragLites-Minimal, Halogenated Fragments Displaying Pharmacophore Doublets. An Efficient Approach to Druggability Assessment and Hit Generation.
J.Med.Chem., 62, 2019
|
|
6Q4J
 
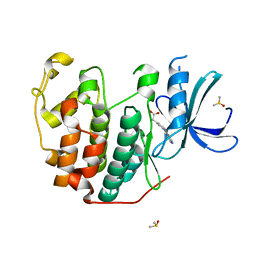 | | CDK2 in complex with FragLite34 | | 分子名称: | 2-[3-(pyrimidin-4-ylamino)phenyl]ethanoic acid, Cyclin-dependent kinase 2, DIMETHYL SULFOXIDE | | 著者 | Wood, D.J, Martin, M.P, Noble, M.E.M. | | 登録日 | 2018-12-05 | | 公開日 | 2019-03-20 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.05 Å) | | 主引用文献 | FragLites-Minimal, Halogenated Fragments Displaying Pharmacophore Doublets. An Efficient Approach to Druggability Assessment and Hit Generation.
J.Med.Chem., 62, 2019
|
|
1TD1
 
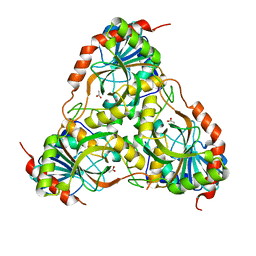 | | Crystal Structure of the Purine Nucleoside Phosphorylase from Schistosoma mansoni in complex with acetate | | 分子名称: | ACETATE ION, purine-nucleoside phosphorylase | | 著者 | Pereira, H.D, Franco, G.R, Cleasby, A, Garratt, R.C. | | 登録日 | 2004-05-21 | | 公開日 | 2005-05-24 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structures for the Potential Drug Target Purine Nucleoside Phosphorylase from Schistosoma mansoni Causal Agent of Schistosomiasis.
J.Mol.Biol., 353, 2005
|
|
3TFX
 
 | | Crystal structure of Orotidine 5'-phosphate decarboxylase from Lactobacillus acidophilus | | 分子名称: | Orotidine 5'-phosphate decarboxylase | | 著者 | Satyanarayana, L, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, LaFleur, J, Seidel, R, Villigas, G, Zencheck, W, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | 登録日 | 2011-08-16 | | 公開日 | 2011-09-28 | | 実験手法 | X-RAY DIFFRACTION (2.19 Å) | | 主引用文献 | Crystal structure of Orotidine 5'-phosphate decarboxylase from Lactobacillus acidophilus
To be Published
|
|
3E9J
 
 | |
8Q52
 
 | | A PBP-like protein built from fragments of different folds | | 分子名称: | Leucine-specific-binding protein,Chemotaxis protein CheY, SULFATE ION | | 著者 | Shanmugaratnam, S, Toledo-Patino, S, Goetz, S.K, Farias-Rico, J.A, Hocker, B. | | 登録日 | 2023-08-08 | | 公開日 | 2024-04-10 | | 最終更新日 | 2024-06-19 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | Molecular handcraft of a well-folded protein chimera.
Febs Lett., 598, 2024
|
|
3E9R
 
 | | Crystal structure of purine nucleoside phosphorylase from Schistosoma mansoni in complex with adenine | | 分子名称: | ACETATE ION, ADENINE, DIMETHYL SULFOXIDE, ... | | 著者 | Pereira, H.M, Rezende, M.M, Oliva, G, Garratt, R.C. | | 登録日 | 2008-08-23 | | 公開日 | 2009-09-01 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Adenosine binding to low-molecular-weight purine nucleoside phosphorylase: the structural basis for recognition based on its complex with the enzyme from Schistosoma mansoni.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
5JRF
 
 | | Crystal structure of the light-driven sodium pump KR2 bound with iodide ions | | 分子名称: | EICOSANE, IODIDE ION, Sodium pumping rhodopsin | | 著者 | Melnikov, I, Polovinkin, V, Kovalev, K, Shevchenko, V, Gushchin, I, Popov, A, Gordeliy, V. | | 登録日 | 2016-05-06 | | 公開日 | 2017-05-31 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Fast iodide-SAD phasing for high-throughput membrane protein structure determination.
Sci Adv, 3, 2017
|
|
5K2D
 
 | | 1.9A angstrom A2a adenosine receptor structure with MR phasing using XFEL data | | 分子名称: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 4-{2-[(7-amino-2-furan-2-yl[1,2,4]triazolo[1,5-a][1,3,5]triazin-5-yl)amino]ethyl}phenol, ... | | 著者 | Batyuk, A, Galli, L, Ishchenko, A, Han, G.W, Gati, C, Popov, P, Lee, M.-Y, Stauch, B, White, T.A, Barty, A, Aquila, A, Hunter, M.S, Liang, M, Boutet, S, Pu, M, Liu, Z.-J, Nelson, G, James, D, Li, C, Zhao, Y, Spence, J.C.H, Liu, W, Fromme, P, Katritch, V, Weierstall, U, Stevens, R.C, Cherezov, V, GPCR Network (GPCR) | | 登録日 | 2016-05-18 | | 公開日 | 2016-09-21 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Native phasing of x-ray free-electron laser data for a G protein-coupled receptor.
Sci Adv, 2, 2016
|
|
5K3V
 
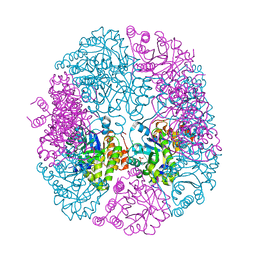 | | apo-PDX1.3 (Arabidopsis) | | 分子名称: | 1,2-ETHANEDIOL, CHLORIDE ION, Pyridoxal 5'-phosphate synthase subunit PDX1.3, ... | | 著者 | Robinson, G.C, Kaufmann, M, Roux, C, Fitzpatrick, T.B. | | 登録日 | 2016-05-20 | | 公開日 | 2016-10-05 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structural definition of the lysine swing in Arabidopsis thaliana PDX1: Intermediate channeling facilitating vitamin B6 biosynthesis.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5K47
 
 | | CryoEM structure of the human Polycystin-2/PKD2 TRP channel | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Polycystin-2 | | 著者 | Pike, A.C.W, Grieben, M, Shintre, C.A, Tessitore, A, Shrestha, L, Mukhopadhyay, S, Mahajan, P, Chalk, R, Burgess-Brown, N.A, Huiskonen, J.T, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Carpenter, E.P, Structural Genomics Consortium (SGC) | | 登録日 | 2016-05-20 | | 公開日 | 2016-08-24 | | 最終更新日 | 2020-10-21 | | 実験手法 | ELECTRON MICROSCOPY (4.2 Å) | | 主引用文献 | Structure of the polycystic kidney disease TRP channel Polycystin-2 (PC2).
Nat. Struct. Mol. Biol., 24, 2017
|
|
5D9A
 
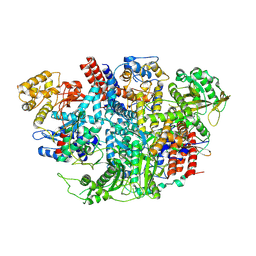 | | Influenza C Virus RNA-dependent RNA Polymerase - Space group P212121 | | 分子名称: | Polymerase acidic protein, Polymerase basic protein 2, RNA-directed RNA polymerase catalytic subunit | | 著者 | Hengrung, N, El Omari, K, Serna Martin, I, Vreede, F.T, Cusack, S, Rambo, R.P, Vonrhein, C, Bricogne, G, Stuart, D.I, Grimes, J.M, Fodor, E. | | 登録日 | 2015-08-18 | | 公開日 | 2015-10-21 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (4.3 Å) | | 主引用文献 | Crystal structure of the RNA-dependent RNA polymerase from influenza C virus.
Nature, 527, 2015
|
|
5K2C
 
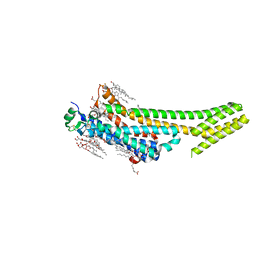 | | 1.9 angstrom A2a adenosine receptor structure with sulfur SAD phasing and phase extension using XFEL data | | 分子名称: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 4-{2-[(7-amino-2-furan-2-yl[1,2,4]triazolo[1,5-a][1,3,5]triazin-5-yl)amino]ethyl}phenol, Adenosine receptor A2a/Soluble cytochrome b562 chimera, ... | | 著者 | Batyuk, A, Galli, L, Ishchenko, A, Han, G.W, Gati, C, Popov, P, Lee, M.-Y, Stauch, B, White, T.A, Barty, A, Aquila, A, Hunter, M.S, Liang, M, Boutet, S, Pu, M, Liu, Z.-J, Nelson, G, James, D, Li, C, Zhao, Y, Spence, J.C.H, Liu, W, Fromme, P, Katritch, V, Weierstall, U, Stevens, R.C, Cherezov, V, GPCR Network (GPCR) | | 登録日 | 2016-05-18 | | 公開日 | 2016-09-21 | | 最終更新日 | 2018-11-28 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Native phasing of x-ray free-electron laser data for a G protein-coupled receptor.
Sci Adv, 2, 2016
|
|
5KE0
 
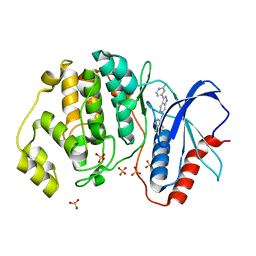 | | Discovery of 1-1H-Pyrazolo 4,3-c pyridine-6-yl urea Inhibitors of Extracellular Signal Regulated Kinase ERK for the Treatment of Cancers | | 分子名称: | 1-[3-(2-methylpyridin-4-yl)-1~{H}-pyrazolo[4,3-c]pyridin-6-yl]-3-(phenylmethyl)urea, Mitogen-activated protein kinase 1, SULFATE ION | | 著者 | Hruza, A, Lim, J. | | 登録日 | 2016-06-09 | | 公開日 | 2016-07-06 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.68 Å) | | 主引用文献 | Discovery of 1-(1H-Pyrazolo[4,3-c]pyridin-6-yl)urea Inhibitors of Extracellular Signal-Regulated Kinase (ERK) for the Treatment of Cancers.
J.Med.Chem., 59, 2016
|
|
5D6Q
 
 | | Crystal structure of the ATP binding domain of S. aureus GyrB complexed with a ligand | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1-ethyl-3-{4-[(E)-2-(pyridin-3-yl)ethenyl]-5-(1H-pyrrol-2-yl)-1,3-thiazol-2-yl}urea, DNA gyrase subunit B, ... | | 著者 | Zhang, J, Yang, Q, Cross, J.B, Romero, J.A.C, Ryan, M.D, Lippa, B, Dolle, R.E, Andersen, O.A, Barker, J, Cheng, R.K, Kahmann, J, Felicetti, B, Wood, M, Scheich, C. | | 登録日 | 2015-08-12 | | 公開日 | 2015-11-25 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Discovery of Azaindole Ureas as a Novel Class of Bacterial Gyrase B Inhibitors.
J.Med.Chem., 58, 2015
|
|
5JSI
 
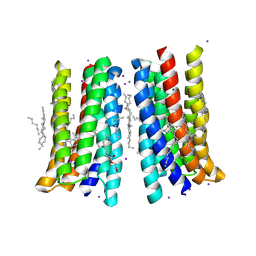 | | Structure of membrane protein | | 分子名称: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Bacteriorhodopsin, EICOSANE, ... | | 著者 | Melnikov, I, Polovinkin, V, Kovalev, K, Shevchenko, V, Gushchin, I, Popov, A, Gordeliy, V. | | 登録日 | 2016-05-08 | | 公開日 | 2017-05-31 | | 最終更新日 | 2023-03-08 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Fast iodide-SAD phasing for high-throughput membrane protein structure determination.
Sci Adv, 3, 2017
|
|
5K2Z
 
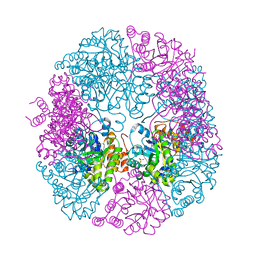 | | PDX1.3-adduct (Arabidopsis) | | 分子名称: | 1,2-ETHANEDIOL, 2-azanylpenta-1,4-dien-3-one, CHLORIDE ION, ... | | 著者 | Robinson, G.C, Kaufmann, M, Roux, C, Fitzpatrick, T.B. | | 登録日 | 2016-05-19 | | 公開日 | 2016-10-05 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structural definition of the lysine swing in Arabidopsis thaliana PDX1: Intermediate channeling facilitating vitamin B6 biosynthesis.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5K2A
 
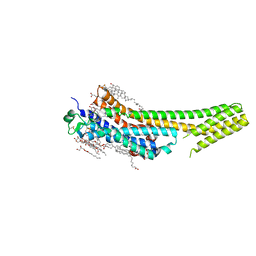 | | 2.5 angstrom A2a adenosine receptor structure with sulfur SAD phasing using XFEL data | | 分子名称: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 4-{2-[(7-amino-2-furan-2-yl[1,2,4]triazolo[1,5-a][1,3,5]triazin-5-yl)amino]ethyl}phenol, Adenosine receptor A2a/Soluble cytochrome b562 chimera, ... | | 著者 | Batyuk, A, Galli, L, Ishchenko, A, Han, G.W, Gati, C, Popov, P, Lee, M.-Y, Stauch, B, White, T.A, Barty, A, Aquila, A, Hunter, M.S, Liang, M, Boutet, S, Pu, M, Liu, Z.-J, Nelson, G, James, D, Li, C, Zhao, Y, Spence, J.C.H, Liu, W, Fromme, P, Katritch, V, Weierstall, U, Stevens, R.C, Cherezov, V, GPCR Network (GPCR) | | 登録日 | 2016-05-18 | | 公開日 | 2016-09-21 | | 最終更新日 | 2018-11-28 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Native phasing of x-ray free-electron laser data for a G protein-coupled receptor.
Sci Adv, 2, 2016
|
|
5CTC
 
 | |
7B2I
 
 | | Heterodimeric tRNA-Guanine Transglycosylase from mouse | | 分子名称: | CITRIC ACID, DI(HYDROXYETHYL)ETHER, HEXAETHYLENE GLYCOL, ... | | 著者 | Sebastiani, M, Heine, A, Reuter, K. | | 登録日 | 2020-11-27 | | 公開日 | 2021-12-08 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Structural and Biochemical Investigation of the Heterodimeric Murine tRNA-Guanine Transglycosylase.
Acs Chem.Biol., 17, 2022
|
|
