4X24
 
 | | Crystal structure of Vibrio cholerae 5'-methylthioadenosine/S-adenosyl homocysteine nucleosidase (MTAN) complexed with methylthio-DADMe-Immucillin-A | | 分子名称: | (3R,4S)-1-[(4-AMINO-5H-PYRROLO[3,2-D]PYRIMIDIN-7-YL)METHYL]-4-[(METHYLSULFANYL)METHYL]PYRROLIDIN-3-OL, 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase, TRIETHYLENE GLYCOL | | 著者 | Cameron, S.A, Thomas, K, Almo, S.C, Schramm, V.L. | | 登録日 | 2014-11-25 | | 公開日 | 2015-08-19 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Active site and remote contributions to catalysis in methylthioadenosine nucleosidases.
Biochemistry, 54, 2015
|
|
3OOH
 
 | | Crystal structure of E. Coli purine nucleoside phosphorylase with PO4 | | 分子名称: | PHOSPHATE ION, Purine nucleoside phosphorylase deoD-type | | 著者 | Mikleusevic, G, Stefanic, Z, Narzyk, M, Wielgus-Kutrowska, B, Bzowska, A, Luic, M. | | 登録日 | 2010-08-31 | | 公開日 | 2011-07-13 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Validation of the catalytic mechanism of Escherichia coli purine nucleoside phosphorylase by structural and kinetic studies.
Biochimie, 93, 2011
|
|
3ONV
 
 | | Crystal structure of E. Coli purine nucleoside phosphorylase with SO4 | | 分子名称: | Purine nucleoside phosphorylase deoD-type, SULFATE ION | | 著者 | Mikleusevic, G, Stefanic, Z, Narzyk, M, Wielgus-Kutrowska, B, Bzowska, A, Luic, M. | | 登録日 | 2010-08-30 | | 公開日 | 2011-07-13 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.893 Å) | | 主引用文献 | Validation of the catalytic mechanism of Escherichia coli purine nucleoside phosphorylase by structural and kinetic studies.
Biochimie, 93, 2011
|
|
3LGS
 
 | |
4DAB
 
 | | Crystal structure of the hexameric purine nucleoside phosphorylase from Bacillus subtilis in complex with hypoxanthine | | 分子名称: | ACETATE ION, CHLORIDE ION, GLYCEROL, ... | | 著者 | Martins, N.H, Giuseppe, P.O, Meza, A.N, Murakami, M.T. | | 登録日 | 2012-01-12 | | 公開日 | 2012-09-26 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Insights into phosphate cooperativity and influence of substrate modifications on binding and catalysis of hexameric purine nucleoside phosphorylases.
Plos One, 7, 2012
|
|
4DAR
 
 | | Crystal structure of the hexameric purine nucleoside phosphorylase from Bacillus subtilis in complex with tubercidin | | 分子名称: | '2-(4-AMINO-PYRROLO[2,3-D]PYRIMIDIN-7-YL)-5-HYDROXYMETHYL-TETRAHYDRO-FURAN-3,4-DIOL, Purine nucleoside phosphorylase deoD-type | | 著者 | Giuseppe, P.O, Martins, N.H, Meza, A.N, Murakami, M.T. | | 登録日 | 2012-01-13 | | 公開日 | 2012-09-26 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (3.15 Å) | | 主引用文献 | Insights into phosphate cooperativity and influence of substrate modifications on binding and catalysis of hexameric purine nucleoside phosphorylases.
Plos One, 7, 2012
|
|
4DAO
 
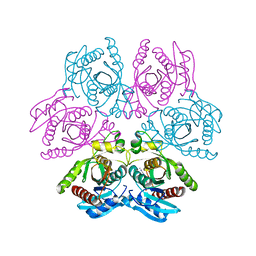 | | Crystal structure of the hexameric purine nucleoside phosphorylase from Bacillus subtilis in complex with adenine | | 分子名称: | ADENINE, GLYCEROL, Purine nucleoside phosphorylase deoD-type | | 著者 | Giuseppe, P.O, Martins, N.H, Meza, A.N, Murakami, M.T. | | 登録日 | 2012-01-13 | | 公開日 | 2012-09-26 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.22 Å) | | 主引用文献 | Insights into phosphate cooperativity and influence of substrate modifications on binding and catalysis of hexameric purine nucleoside phosphorylases.
Plos One, 7, 2012
|
|
4DAE
 
 | | Crystal structure of the hexameric purine nucleoside phosphorylase from Bacillus subtilis in complex with 6-chloroguanosine | | 分子名称: | 6-chloro-9-(beta-D-ribofuranosyl)-9H-purin-2-amine, ACETATE ION, CHLORIDE ION, ... | | 著者 | Martins, N.H, Giuseppe, P.O, Meza, A.N, Murakami, M.T. | | 登録日 | 2012-01-12 | | 公開日 | 2012-09-26 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | Insights into phosphate cooperativity and influence of substrate modifications on binding and catalysis of hexameric purine nucleoside phosphorylases.
Plos One, 7, 2012
|
|
4DAN
 
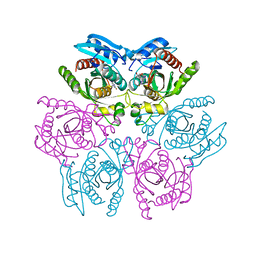 | | Crystal structure of the hexameric purine nucleoside phosphorylase from Bacillus subtilis in complex with 2-fluoroadenosine | | 分子名称: | 2-(6-AMINO-2-FLUORO-PURIN-9-YL)-5-HYDROXYMETHYL-TETRAHYDRO-FURAN-3,4-DIOL, Purine nucleoside phosphorylase deoD-type | | 著者 | Giuseppe, P.O, Martins, N.H, Meza, A.N, Murakami, M.T. | | 登録日 | 2012-01-13 | | 公開日 | 2012-09-26 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.56 Å) | | 主引用文献 | Insights into phosphate cooperativity and influence of substrate modifications on binding and catalysis of hexameric purine nucleoside phosphorylases.
Plos One, 7, 2012
|
|
6FF4
 
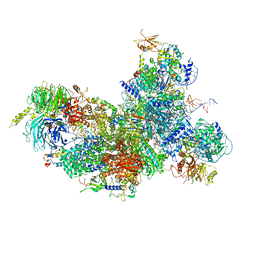 | | human Bact spliceosome core structure | | 分子名称: | 116 kDa U5 small nuclear ribonucleoprotein component, BUD13 homolog, Cell division cycle 5-like protein, ... | | 著者 | Haselbach, D, Komarov, I, Agafonov, D, Hartmuth, K, Graf, B, Kastner, B, Luehrmann, R, Stark, H. | | 登録日 | 2018-01-03 | | 公開日 | 2018-08-29 | | 最終更新日 | 2025-07-02 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Structure and Conformational Dynamics of the Human Spliceosomal BactComplex.
Cell, 172, 2018
|
|
4DA8
 
 | | Crystal structure of the hexameric purine nucleoside phosphorylase from Bacillus subtilis in complex with 8-bromoguanosine | | 分子名称: | 8-bromoguanosine, Purine nucleoside phosphorylase deoD-type | | 著者 | Martins, N.H, Giuseppe, P.O, Meza, A.N, Murakami, M.T. | | 登録日 | 2012-01-12 | | 公開日 | 2012-09-26 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Insights into phosphate cooperativity and influence of substrate modifications on binding and catalysis of hexameric purine nucleoside phosphorylases.
Plos One, 7, 2012
|
|
3IOM
 
 | |
5O9G
 
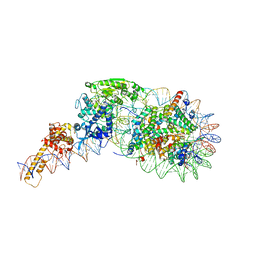 | | Structure of nucleosome-Chd1 complex | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, Chromo domain-containing protein 1, ... | | 著者 | Farnung, L, Vos, S.M, Wigge, C, Cramer, P. | | 登録日 | 2017-06-19 | | 公開日 | 2017-10-11 | | 最終更新日 | 2024-05-15 | | 実験手法 | ELECTRON MICROSCOPY (4.8 Å) | | 主引用文献 | Nucleosome-Chd1 structure and implications for chromatin remodelling.
Nature, 550, 2017
|
|
3IX2
 
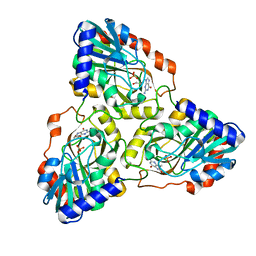 | |
3OOE
 
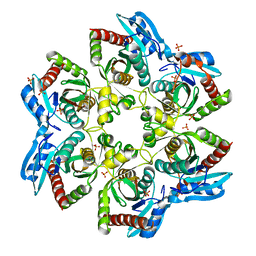 | | Crystal structure of E. Coli purine nucleoside phosphorylase with PO4 | | 分子名称: | PHOSPHATE ION, Purine nucleoside phosphorylase deoD-type | | 著者 | Mikleusevic, G, Stefanic, Z, Narzyk, M, Wielgus-Kutrowska, B, Bzowska, A, Luic, M. | | 登録日 | 2010-08-31 | | 公開日 | 2011-07-13 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Validation of the catalytic mechanism of Escherichia coli purine nucleoside phosphorylase by structural and kinetic studies.
Biochimie, 93, 2011
|
|
4DA7
 
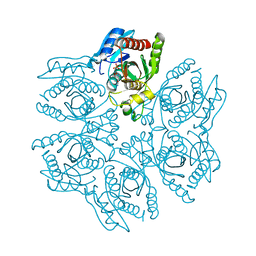 | | Crystal structure of the hexameric purine nucleoside phosphorylase from Bacillus subtilis in complex with aciclovir | | 分子名称: | 9-HYROXYETHOXYMETHYLGUANINE, PHOSPHATE ION, Purine nucleoside phosphorylase deoD-type | | 著者 | Martins, N.H, Giuseppe, P.O, Meza, A.N, Murakami, M.T. | | 登録日 | 2012-01-12 | | 公開日 | 2012-09-26 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Insights into phosphate cooperativity and influence of substrate modifications on binding and catalysis of hexameric purine nucleoside phosphorylases.
Plos One, 7, 2012
|
|
3OPV
 
 | | Crystal structure of E. Coli purine nucleoside phosphorylase Arg24Ala mutant | | 分子名称: | PHOSPHATE ION, Purine nucleoside phosphorylase deoD-type | | 著者 | Mikleusevic, G, Stefanic, Z, Narzyk, M, Wielgus-Kutrowska, B, Bzowska, A, Luic, M. | | 登録日 | 2010-09-02 | | 公開日 | 2011-07-13 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Validation of the catalytic mechanism of Escherichia coli purine nucleoside phosphorylase by structural and kinetic studies.
Biochimie, 93, 2011
|
|
1G2O
 
 | | CRYSTAL STRUCTURE OF PURINE NUCLEOSIDE PHOSPHORYLASE FROM MYCOBACTERIUM TUBERCULOSIS IN COMPLEX WITH A TRANSITION-STATE INHIBITOR | | 分子名称: | 1,4-DIDEOXY-4-AZA-1-(S)-(9-DEAZAHYPOXANTHIN-9-YL)-D-RIBITOL, PHOSPHATE ION, PURINE NUCLEOSIDE PHOSPHORYLASE | | 著者 | Shi, W, Basso, L.A, Tyler, P.C, Furneaux, R.H, Blanchard, J.S, Almo, S.C, Schramm, V.L. | | 登録日 | 2000-10-20 | | 公開日 | 2001-08-01 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Structures of purine nucleoside phosphorylase from Mycobacterium tuberculosis in complexes with immucillin-H and its pieces.
Biochemistry, 40, 2001
|
|
3MB8
 
 | | Crystal structure of purine nucleoside phosphorylase from toxoplasma gondii in complex with immucillin-H | | 分子名称: | 1,4-DIDEOXY-4-AZA-1-(S)-(9-DEAZAHYPOXANTHIN-9-YL)-D-RIBITOL, GLYCEROL, PHOSPHATE ION, ... | | 著者 | Ho, M, Almo, S.C, Schramm, V.L. | | 登録日 | 2010-03-25 | | 公開日 | 2011-04-06 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Inhibition and Structure of Toxoplasma gondii Purine Nucleoside Phosphorylase.
Eukaryot Cell, 13, 2014
|
|
5MX8
 
 | |
4ANE
 
 | | R80N MUTANT OF NUCLEOSIDE DIPHOSPHATE KINASE FROM MYCOBACTERIUM TUBERCULOSIS | | 分子名称: | CITRIC ACID, NUCLEOSIDE DIPHOSPHATE KINASE | | 著者 | Georgescauld, F, Moynie, L, Habersetzer, J, Lascu, I, Dautant, A. | | 登録日 | 2012-03-16 | | 公開日 | 2013-03-13 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structure of Mycobacterium Tuberculosis Nucleoside Diphosphate Kinase R80N Mutant in Complex with Citrate
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3ZTP
 
 | | Orthorhombic crystal form P21212 of the Aquifex aeolicus nucleoside diphosphate kinase | | 分子名称: | GLYCEROL, NUCLEOSIDE DIPHOSPHATE KINASE, SULFATE ION | | 著者 | Boissier, F, Georgescauld, F, Moynie, L, Dupuy, J.-W, Sarger, C, Podar, M, Lascu, I, Giraud, M.-F, Dautant, A. | | 登録日 | 2011-07-12 | | 公開日 | 2012-03-14 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (1.37 Å) | | 主引用文献 | An Intersubunit Disulfide Bridge Stabilizes the Tetrameric Nucleoside Diphosphate Kinase of Aquifex Aeolicus.
Proteins, 80, 2012
|
|
2F64
 
 | | Crystal structure of Nucleoside 2-deoxyribosyltransferase from Trypanosoma brucei at 1.6 A resolution with 1-METHYLQUINOLIN-2(1H)-ONE bound | | 分子名称: | 1-METHYLQUINOLIN-2(1H)-ONE, GLYCEROL, Nucleoside 2-deoxyribosyltransferase, ... | | 著者 | Bosch, J, Robien, M.A, Hol, W.G.J, Structural Genomics of Pathogenic Protozoa Consortium (SGPP) | | 登録日 | 2005-11-28 | | 公開日 | 2005-12-06 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Using fragment cocktail crystallography to assist inhibitor design of Trypanosoma brucei nucleoside 2-deoxyribosyltransferase.
J.Med.Chem., 49, 2006
|
|
5MX4
 
 | |
2F62
 
 | | Crystal structure of Nucleoside 2-deoxyribosyltransferase from Trypanosoma brucei at 1.5 A resolution with (2-ETHYLPHENYL)METHANOL bound | | 分子名称: | (2-ETHYLPHENYL)METHANOL, GLYCEROL, Nucleoside 2-deoxyribosyltransferase, ... | | 著者 | Bosch, J, Robien, M.A, Hol, W.G.J, Structural Genomics of Pathogenic Protozoa Consortium (SGPP) | | 登録日 | 2005-11-28 | | 公開日 | 2005-12-06 | | 最終更新日 | 2024-10-09 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Using fragment cocktail crystallography to assist inhibitor design of Trypanosoma brucei nucleoside 2-deoxyribosyltransferase.
J.Med.Chem., 49, 2006
|
|
