5Q12
 
 | | Ligand binding to FARNESOID-X-RECEPTOR | | 分子名称: | 2-(2,6-difluorophenyl)-N-(2,6-dimethylphenyl)-5-methylimidazo[1,2-a]pyridin-3-amine, Bile acid receptor, COACTIVATOR PEPTIDE SRC-1 HD3 | | 著者 | Rudolph, M.G, Benz, J, Burger, D, Thoma, R, Ruf, A, Joseph, C, Kuhn, B, Shao, C, Yang, H, Burley, S.K. | | 登録日 | 2017-05-31 | | 公開日 | 2017-07-05 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | D3R Grand Challenge 2: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J. Comput. Aided Mol. Des., 32, 2018
|
|
5Q1G
 
 | | Ligand binding to FARNESOID-X-RECEPTOR | | 分子名称: | (2E)-N-cyclohexyl-N-(cyclohexylcarbamoyl)-3-(4-fluorophenyl)prop-2-enamide, Bile acid receptor, COACTIVATOR PEPTIDE SRC-1 HD3 | | 著者 | Rudolph, M.G, Benz, J, Burger, D, Thoma, R, Ruf, A, Joseph, C, Kuhn, B, Shao, C, Yang, H, Burley, S.K. | | 登録日 | 2017-05-31 | | 公開日 | 2017-07-05 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | D3R Grand Challenge 2: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J. Comput. Aided Mol. Des., 32, 2018
|
|
5Q0U
 
 | | Ligand binding to FARNESOID-X-RECEPTOR | | 分子名称: | Bile acid receptor, COACTIVATOR PEPTIDE SRC-1 HD3, trans-4-({(2S)-2-[2-(4-chlorophenyl)-5,6-difluoro-1H-benzimidazol-1-yl]-2-cyclohexylacetyl}amino)cyclohexyl hydrogen sulfate | | 著者 | Rudolph, M.G, Benz, J, Burger, D, Thoma, R, Ruf, A, Joseph, C, Kuhn, B, Shao, C, Yang, H, Burley, S.K. | | 登録日 | 2017-05-31 | | 公開日 | 2017-07-05 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | D3R Grand Challenge 2: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J. Comput. Aided Mol. Des., 32, 2018
|
|
5Q1C
 
 | | Ligand binding to FARNESOID-X-RECEPTOR | | 分子名称: | (2S)-2-cyclohexyl-2-[2-(2,6-dimethoxypyridin-3-yl)-5,6-difluoro-1H-benzimidazol-1-yl]-N-(trans-4-hydroxycyclohexyl)acetamide, Bile acid receptor, COACTIVATOR PEPTIDE SRC-1 HD3 | | 著者 | Rudolph, M.G, Benz, J, Burger, D, Thoma, R, Ruf, A, Joseph, C, Kuhn, B, Shao, C, Yang, H, Burley, S.K. | | 登録日 | 2017-05-31 | | 公開日 | 2017-07-05 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | D3R Grand Challenge 2: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J. Comput. Aided Mol. Des., 32, 2018
|
|
5Q0O
 
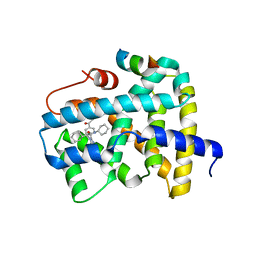 | | Ligand binding to FARNESOID-X-RECEPTOR | | 分子名称: | (2S)-2-{2-[4-(benzenecarbonyl)phenyl]-1H-benzimidazol-1-yl}-N,2-dicyclohexylacetamide, Bile acid receptor, CHLORIDE ION, ... | | 著者 | Rudolph, M.G, Benz, J, Burger, D, Thoma, R, Ruf, A, Joseph, C, Kuhn, B, Shao, C, Yang, H, Burley, S.K. | | 登録日 | 2017-05-31 | | 公開日 | 2017-07-05 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | D3R Grand Challenge 2: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J. Comput. Aided Mol. Des., 32, 2018
|
|
5Q14
 
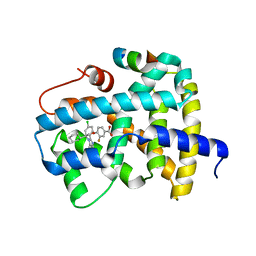 | | Ligand binding to FARNESOID-X-RECEPTOR | | 分子名称: | 4-{(2S)-2-[2-(4-chlorophenyl)-5,6-difluoro-1H-benzimidazol-1-yl]-2-cyclohexylethoxy}-3-fluorobenzoic acid, Bile acid receptor, COACTIVATOR PEPTIDE SRC-1 HD3 | | 著者 | Rudolph, M.G, Benz, J, Burger, D, Thoma, R, Ruf, A, Joseph, C, Kuhn, B, Shao, C, Yang, H, Burley, S.K. | | 登録日 | 2017-05-31 | | 公開日 | 2017-07-05 | | 最終更新日 | 2024-11-13 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | D3R Grand Challenge 2: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J. Comput. Aided Mol. Des., 32, 2018
|
|
7PH1
 
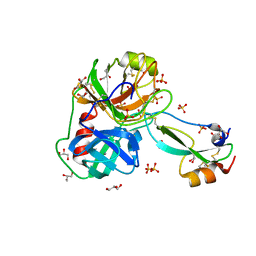 | | Trypsin in complex with BPTI mutant (2S)-2-amino-4-monofluorobutanoic acid | | 分子名称: | CALCIUM ION, Cationic trypsin, GLYCEROL, ... | | 著者 | Dimos, N, Leppkes, J, Koksch, B, Loll, B. | | 登録日 | 2021-08-16 | | 公開日 | 2022-03-30 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.18 Å) | | 主引用文献 | Water Network in the Binding Pocket of Fluorinated BPTI-Trypsin Complexes─Insights from Simulation and Experiment.
J.Phys.Chem.B, 126, 2022
|
|
8AZC
 
 | |
4MLO
 
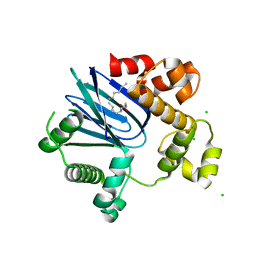 | | 1.65A resolution structure of ToxT from Vibrio cholerae (P21 Form) | | 分子名称: | CHLORIDE ION, PALMITOLEIC ACID, TCP pilus virulence regulatory protein | | 著者 | Lovell, S, Wehmeyer, G, Battaile, K.P, Li, J, Egan, S. | | 登録日 | 2013-09-06 | | 公開日 | 2016-04-20 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | 1.65 angstrom resolution structure of the AraC-family transcriptional activator ToxT from Vibrio cholerae.
Acta Crystallogr F Struct Biol Commun, 72, 2016
|
|
8DFZ
 
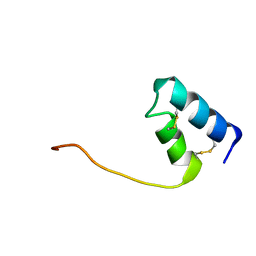 | |
8C3T
 
 | |
4QQK
 
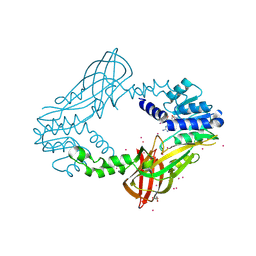 | | Human HMT1 hnRNP methyltransferase-like protein 6 (S. cerevisiae) with GMS | | 分子名称: | (5S)-5-{[(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl}-N~6~-carbamimidoyl-L-lysine, GLYCEROL, Protein arginine N-methyltransferase 6, ... | | 著者 | Dong, A, Zeng, H, He, H, Wernimont, A, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Brown, P.J, Min, J, Luo, M, Wu, H, Structural Genomics Consortium (SGC) | | 登録日 | 2014-06-27 | | 公開日 | 2014-07-16 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (1.88 Å) | | 主引用文献 | Structural basis of arginine asymmetrical dimethylation by PRMT6.
Biochem. J., 473, 2016
|
|
7OIH
 
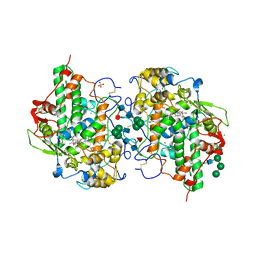 | | Glycosylation in the crystal structure of neutrophil myeloperoxidase | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Krawczyk, L, Semwal, S, Bouckaert, J. | | 登録日 | 2021-05-11 | | 公開日 | 2022-08-24 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (2.603 Å) | | 主引用文献 | Native glycosylation and binding of the antidepressant paroxetine in a low-resolution crystal structure of human myeloperoxidase.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7P4Z
 
 | |
8R8E
 
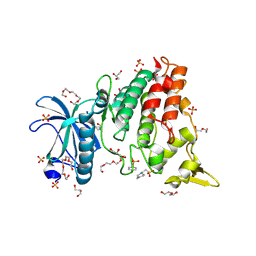 | | DYRK1a in Complex with 2-Cyclopentyl-7-iodo-1H-indole-3-carbonitrile | | 分子名称: | 1,2-ETHANEDIOL, 2-cyclopentyl-7-iodanyl-1~{H}-indole-3-carbonitrile, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | 著者 | Stahlecker, J, Dammann, M, Stehle, T, Boeckler, F.M. | | 登録日 | 2023-11-29 | | 公開日 | 2024-09-25 | | 最終更新日 | 2024-12-18 | | 実験手法 | X-RAY DIFFRACTION (2.22 Å) | | 主引用文献 | Halogen Bonding on Water─A Drop in the Ocean?
J Chem Theory Comput, 20, 2024
|
|
4YE7
 
 | |
7ZBF
 
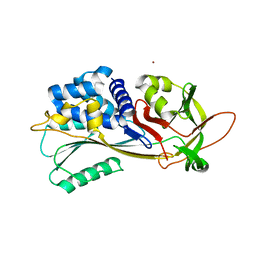 | | Crystal structure of native Iripin-4 serpin from tick Ixodes ricinus | | 分子名称: | Iripin-4 serpin, NICKEL (II) ION | | 著者 | Kascakova, B, Kuta Smatanova, I, Chmelar, J, Prudnikova, T. | | 登録日 | 2022-03-23 | | 公開日 | 2023-04-05 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Conformational transition of the Ixodes ricinus salivary serpin Iripin-4.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
7PJD
 
 | |
5DLP
 
 | | Acetylcholinesterase Methylene Blue no PEG | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 3,7-BIS(DIMETHYLAMINO)PHENOTHIAZIN-5-IUM, Acetylcholinesterase, ... | | 著者 | Dym, O. | | 登録日 | 2015-09-07 | | 公開日 | 2016-03-30 | | 最終更新日 | 2024-11-13 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | The impact of crystallization conditions on structure-based drug design: A case study on the methylene blue/acetylcholinesterase complex.
Protein Sci., 25, 2016
|
|
6XP4
 
 | |
6XPS
 
 | |
6XPW
 
 | |
5L88
 
 | | AFAMIN ANTIBODY FRAGMENT, N14 FAB, L1- GLYCOSILATED, CRYSTAL FORM I, non-parsimonious model | | 分子名称: | 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, Anti-afamin antibody N14, Fab fragment, ... | | 著者 | Rupp, B, Naschberger, A. | | 登録日 | 2016-06-07 | | 公開日 | 2016-08-03 | | 最終更新日 | 2024-11-20 | | 実験手法 | X-RAY DIFFRACTION (1.88 Å) | | 主引用文献 | The N14 anti-afamin antibody Fab: a rare VL1 CDR glycosylation, crystallographic re-sequencing, molecular plasticity and conservative versus enthusiastic modelling.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
6Y9S
 
 | | Crystal structure of GSK-3b in complex with the imidazo[1,5-a]pyridine-3-carboxamide inhibitor 16 | | 分子名称: | ACETATE ION, Glycogen synthase kinase-3 beta, ~{N}-(oxan-4-ylmethyl)-6-(5-propan-2-yloxypyridin-3-yl)imidazo[1,5-a]pyridine-3-carboxamide | | 著者 | Krapp, S, Griessner, A, Blaesse, M, Buonfiglio, R, Ombrato, R. | | 登録日 | 2020-03-10 | | 公開日 | 2020-05-20 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (2.03 Å) | | 主引用文献 | Discovery of Novel Imidazopyridine GSK-3 beta Inhibitors Supported by Computational Approaches.
Molecules, 25, 2020
|
|
6XP7
 
 | |
