5N4K
 
 | | N-terminal domain of a human Coronavirus NL63 nucleocapsid protein | | 分子名称: | Nucleoprotein, SULFATE ION | | 著者 | Zdzalik, M, Szelazek, B, Kabala, W, Golik, P, Burmistrz, M, Florek, D, Kus, K, Pyrc, K, Dubin, G. | | 登録日 | 2017-02-10 | | 公開日 | 2017-02-22 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.49 Å) | | 主引用文献 | Structural Characterization of Human Coronavirus NL63 N Protein.
J. Virol., 91, 2017
|
|
2GMS
 
 | | E coli GDP-4-keto-6-deoxy-D-mannose-3-dehydratase with bound hydrated PLP | | 分子名称: | MAGNESIUM ION, Putative pyridoxamine 5-phosphate-dependent dehydrase, Wbdk, ... | | 著者 | Cook, P.D, Thoden, J.B, Holden, H.M. | | 登録日 | 2006-04-07 | | 公開日 | 2006-09-05 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | The structure of GDP-4-keto-6-deoxy-D-mannose-3-dehydratase: a unique coenzyme B6-dependent enzyme.
Protein Sci., 15, 2006
|
|
3J07
 
 | | Model of a 24mer alphaB-crystallin multimer | | 分子名称: | Alpha-crystallin B chain | | 著者 | Jehle, S, Vollmar, B, Bardiaux, B, Dove, K.K, Rajagopal, P, Gonen, T, Oschkinat, H, Klevit, R.E. | | 登録日 | 2011-04-27 | | 公開日 | 2016-01-20 | | 最終更新日 | 2024-05-01 | | 実験手法 | ELECTRON MICROSCOPY (20 Å), SOLID-STATE NMR, SOLUTION SCATTERING | | 主引用文献 | N-terminal domain of {alpha}B-crystallin provides a conformational switch for multimerization and structural heterogeneity.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
5N1O
 
 | |
2H68
 
 | | Histone H3 recognition and presentation by the WDR5 module of the MLL1 complex | | 分子名称: | WD-repeat protein 5 | | 著者 | Ruthenburg, A.J, Wang, W.-K, Graybosch, D.M, Li, H, Allis, C.D, Patel, D.J, Verdine, G.L. | | 登録日 | 2006-05-30 | | 公開日 | 2006-07-04 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.79 Å) | | 主引用文献 | Histone H3 recognition and presentation by the WDR5 module of the MLL1 complex.
Nat.Struct.Mol.Biol., 13, 2006
|
|
5MZY
 
 | | Crystal structure of the decarboxylase AibA/AibB in complex with a possible transition state analog | | 分子名称: | (1~{R},2~{S})-2-methylcyclohexane-1-carboxylic acid, ACETATE ION, GLYCEROL, ... | | 著者 | Bock, T, Luxenburger, E, Hoffmann, J, Schuetza, V, Feiler, C, Mueller, R, Blankenfeldt, W. | | 登録日 | 2017-02-02 | | 公開日 | 2017-05-31 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | AibA/AibB Induces an Intramolecular Decarboxylation in Isovalerate Biosynthesis by Myxococcus xanthus.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
2H6K
 
 | | Histone H3 recognition and presentation by the WDR5 module of the MLL1 complex | | 分子名称: | Histone H3 K4-Me 9-residue peptide, WD-repeat protein 5 | | 著者 | Ruthenburg, A.J, Wang, W.-K, Graybosch, D.M, Li, H, Allis, C.D, Patel, D.J, Verdine, G.L. | | 登録日 | 2006-05-31 | | 公開日 | 2006-07-04 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.89 Å) | | 主引用文献 | Histone H3 recognition and presentation by the WDR5 module of the MLL1 complex.
Nat.Struct.Mol.Biol., 13, 2006
|
|
7GLQ
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with PET-UNK-8df914d1-2 (Mpro-P2007) | | 分子名称: | 2-(3-chlorophenyl)-N-[(4R)-imidazo[1,2-a]pyridin-3-yl]acetamide, 3C-like proteinase, CHLORIDE ION, ... | | 著者 | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | 登録日 | 2023-08-11 | | 公開日 | 2023-11-08 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (2.063 Å) | | 主引用文献 | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
5N32
 
 | |
5N3J
 
 | | cAMP-dependent Protein Kinase A from Cricetulus griseus in complex with fragment like molecule 4-Nitrobenzoic acid | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, 4-NITROBENZOIC ACID, DIMETHYL SULFOXIDE, ... | | 著者 | Siefker, C, Heine, A, Klebe, G. | | 登録日 | 2017-02-08 | | 公開日 | 2018-02-28 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.12 Å) | | 主引用文献 | Fragment Binding to Kinase Hinge: If Charge Distribution and Local pK a Shifts Mislead Popular Bioisosterism Concepts.
Angew.Chem.Int.Ed.Engl., 2020
|
|
2NV2
 
 | | Structure of the PLP synthase complex Pdx1/2 (YaaD/E) from Bacillus subtilis | | 分子名称: | 1,2-ETHANEDIOL, CHLORIDE ION, GLUTAMINE, ... | | 著者 | Strohmeier, M, Tews, I, Sinning, I. | | 登録日 | 2006-11-10 | | 公開日 | 2006-12-05 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.12 Å) | | 主引用文献 | Structure of a bacterial pyridoxal 5'-phosphate synthase complex
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
5N1K
 
 | |
3IQH
 
 | |
7ET5
 
 | | Crystal structure of Arabidopsis TEM1 AP2 domain | | 分子名称: | AP2/ERF and B3 domain-containing transcription repressor TEM1, SULFATE ION | | 著者 | Hu, H, Du, J. | | 登録日 | 2021-05-12 | | 公開日 | 2021-09-15 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.052 Å) | | 主引用文献 | TEM1 combinatorially binds to FLOWERING LOCUS T and recruits a Polycomb factor to repress the floral transition in Arabidopsis.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
3BG5
 
 | | Crystal Structure of Staphylococcus Aureus Pyruvate Carboxylase | | 分子名称: | 5-(HEXAHYDRO-2-OXO-1H-THIENO[3,4-D]IMIDAZOL-6-YL)PENTANAL, ADENOSINE-5'-TRIPHOSPHATE, MANGANESE (II) ION, ... | | 著者 | Xiang, S, Tong, L. | | 登録日 | 2007-11-26 | | 公開日 | 2008-02-26 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Crystal structures of human and Staphylococcus aureus pyruvate carboxylase and molecular insights into the carboxyltransfer reaction.
Nat.Struct.Mol.Biol., 15, 2008
|
|
5N1T
 
 | | Crystal structure of complex between flavocytochrome c and copper chaperone CopC from T. paradoxus | | 分子名称: | COPPER (II) ION, CopC, Cytochrome C, ... | | 著者 | Osipov, E.M, Lilina, A.V, Tikhonova, T.V, Tsallagov, S.I, Popov, V.O. | | 登録日 | 2017-02-06 | | 公開日 | 2018-02-28 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Structure of the flavocytochrome c sulfide dehydrogenase associated with the copper-binding protein CopC from the haloalkaliphilic sulfur-oxidizing bacterium Thioalkalivibrio paradoxusARh 1.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
2GSY
 
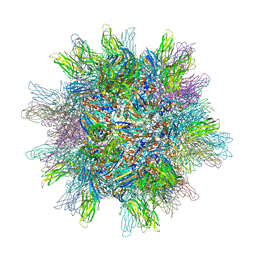 | | The 2.6A structure of Infectious Bursal Virus Derived T=1 Particles | | 分子名称: | CALCIUM ION, polyprotein | | 著者 | Garriga, D, Querol-Audi, J, Abaitua, F, Saugar, I, Pous, J, Verdaguer, N, Caston, J.R, Rodriguez, J.F. | | 登録日 | 2006-04-27 | | 公開日 | 2006-07-18 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | The 2.6-angstrom structure of infectious bursal disease virus-derived t=1 particles reveals new stabilizing elements of the virus capsid.
J.Virol., 80, 2006
|
|
7ET6
 
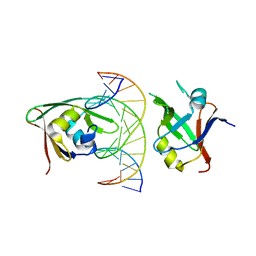 | | Crystal structure of Arabidopsis TEM1 B3-DNA complex | | 分子名称: | AP2/ERF and B3 domain-containing transcription repressor TEM1, FT-RY14-F, FT-RY14-R | | 著者 | Hu, H, Du, J. | | 登録日 | 2021-05-12 | | 公開日 | 2021-09-15 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | TEM1 combinatorially binds to FLOWERING LOCUS T and recruits a Polycomb factor to repress the floral transition in Arabidopsis.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
5N3O
 
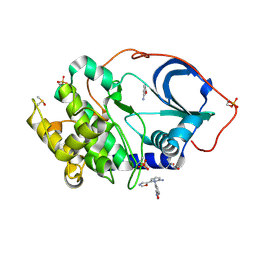 | | cAMP-dependent Protein Kinase A from Cricetulus griseus in complex with fragment like molecule 3-(1,3-oxazol-5-yl)aniline | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, 3-(1,3-oxazol-5-yl)aniline, DIMETHYL SULFOXIDE, ... | | 著者 | Siefker, C, Heine, A, Klebe, G. | | 登録日 | 2017-02-08 | | 公開日 | 2018-02-28 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.32 Å) | | 主引用文献 | A crystallographic fragment study with cAMP-dependent protein kinase A
To Be Published
|
|
7ET4
 
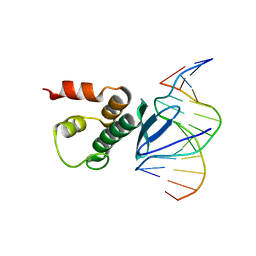 | | Crystal structure of Arabidopsis TEM1 AP2 domain | | 分子名称: | AP2/ERF and B3 domain-containing transcription repressor TEM1, DNA (12-mer) | | 著者 | Hu, H, Du, J. | | 登録日 | 2021-05-12 | | 公開日 | 2021-09-15 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | TEM1 combinatorially binds to FLOWERING LOCUS T and recruits a Polycomb factor to repress the floral transition in Arabidopsis.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7EY9
 
 | | tail proteins | | 分子名称: | Tail fiber protein, Tail tubular protein gp11, Tail tubular protein gp12 | | 著者 | Liu, H.R, Chen, W.Y. | | 登録日 | 2021-05-30 | | 公開日 | 2021-09-22 | | 最終更新日 | 2024-06-12 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Structural changes in bacteriophage T7 upon receptor-induced genome ejection.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
5N6F
 
 | | Crystal structure of TGT in complex with guanine fragment | | 分子名称: | DI(HYDROXYETHYL)ETHER, DIMETHYL SULFOXIDE, GUANINE, ... | | 著者 | Hassaan, E, Heine, A, Klebe, G. | | 登録日 | 2017-02-15 | | 公開日 | 2018-03-07 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.11909521 Å) | | 主引用文献 | Fragments as Novel Starting Points for tRNA-Guanine Transglycosylase Inhibitors Found by Alternative Screening Strategies.
Chemmedchem, 15, 2020
|
|
3IWR
 
 | | Crystal structure of class I chitinase from Oryza sativa L. japonica | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Chitinase | | 著者 | Kezuka, Y, Watanabe, T, Nonaka, T. | | 登録日 | 2009-09-03 | | 公開日 | 2010-04-21 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.57 Å) | | 主引用文献 | Structure of full-length class I chitinase from rice revealed by X-ray crystallography and small-angle X-ray scattering.
Proteins, 78, 2010
|
|
7F26
 
 | | Crystal structure of lysozyme | | 分子名称: | Lysozyme C | | 著者 | Liang, M. | | 登録日 | 2021-06-10 | | 公開日 | 2021-09-15 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Novel combined crystallization plate for high-throughput crystal screening and in situ data collection at a crystallography beamline.
Acta Crystallogr.,Sect.F, 77, 2021
|
|
3IS8
 
 | | Structure of mineralized Bfrb soaked with FeSO4 from Pseudomonas aeruginosa to 2.25A Resolution | | 分子名称: | Bacterioferritin, FE (II) ION, POTASSIUM ION, ... | | 著者 | Lovell, S, Weeratunga, S.K, Battaile, K.P, Rivera, M. | | 登録日 | 2009-08-25 | | 公開日 | 2010-02-02 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Structural Studies of Bacterioferritin B from Pseudomonas aeruginosa Suggest a Gating Mechanism for Iron Uptake via the Ferroxidase Center
Biochemistry, 49, 2010
|
|
