4J6X
 
 | |
2FR3
 
 | |
2FS7
 
 | |
4J8O
 
 | | SET7/9 in complex with TAF10K189A peptide and AdoHcy | | 分子名称: | Histone-lysine N-methyltransferase SETD7, S-ADENOSYL-L-HOMOCYSTEINE, Transcription initiation factor TFIID subunit 10 | | 著者 | Horowitz, S, Trievel, R.C. | | 登録日 | 2013-02-14 | | 公開日 | 2014-01-08 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.63 Å) | | 主引用文献 | Conservation and functional importance of carbon-oxygen hydrogen bonding in AdoMet-dependent methyltransferases.
J.Am.Chem.Soc., 135, 2013
|
|
2YY0
 
 | | Crystal Structure of MS0802, c-Myc-1 binding protein domain from Homo sapiens | | 分子名称: | C-Myc-binding protein | | 著者 | Xie, Y, Wang, H, Ihsanawati, K.T, Kishishita, S, Takemoto, C, Shirozu, M, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2007-04-27 | | 公開日 | 2008-04-29 | | 最終更新日 | 2024-06-26 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | crystal structure of c-Myc-1 binding protein domain from Homo sapiens
To be Published
|
|
2FEX
 
 | | The Crystal Structure of DJ-1 Superfamily Protein Atu0886 from Agrobacterium tumefaciens | | 分子名称: | GLYCEROL, SULFATE ION, conserved hypothetical protein | | 著者 | Cymborowski, M.T, Wang, S, Chruszcz, M, Shumilin, I, Gu, J, Xu, X, Edwards, A.M, Savchenko, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2005-12-16 | | 公開日 | 2006-01-31 | | 最終更新日 | 2022-04-13 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | The Crystal Structure of DJ-1 Superfamily Protein Atu0886 from Agrobacterium tumefaciens
To be Published
|
|
2YHT
 
 | |
4NYA
 
 | | Crystal structure of the E. coli thiM riboswitch in complex with 5-(azidomethyl)-2-methylpyrimidin-4-amine | | 分子名称: | 5-(azidomethyl)-2-methylpyrimidin-4-amine, MAGNESIUM ION, thiM TPP riboswitch | | 著者 | Warner, K.D, Homan, P, Weeks, K.M, Smith, A.G, Abell, C, Ferre-D'Amare, A.R. | | 登録日 | 2013-12-10 | | 公開日 | 2014-06-04 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.65 Å) | | 主引用文献 | Validating Fragment-Based Drug Discovery for Biological RNAs: Lead Fragments Bind and Remodel the TPP Riboswitch Specifically.
Chem.Biol., 21, 2014
|
|
2YC0
 
 | | FACTOR INHIBITING HIF-1 ALPHA IN COMPLEX WITH R-2-HYDROXYGLUTARATE | | 分子名称: | (2R)-2-hydroxypentanedioic acid, FE (II) ION, GLYCEROL, ... | | 著者 | Chowdhury, R, Clifton, I.J, Schofield, C.J. | | 登録日 | 2011-03-10 | | 公開日 | 2011-03-30 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | The oncometabolite 2-hydroxyglutarate inhibits histone lysine demethylases.
EMBO Rep., 12, 2011
|
|
2YF3
 
 | | Crystal structure of DR2231, the MazG-like protein from Deinococcus radiodurans, complex with manganese | | 分子名称: | GLYCEROL, MANGANESE (II) ION, MAZG-LIKE NUCLEOSIDE TRIPHOSPHATE PYROPHOSPHOHYDROLASE, ... | | 著者 | Goncalves, A.M.D, deSanctis, D, McSweeney, S.M. | | 登録日 | 2011-04-01 | | 公開日 | 2011-07-06 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural and Functional Insights Into Dr2231 Protein, the Mazg-Like Nucleoside Triphosphate Pyrophosphohydrolase from Deinococcus Radiodurans.
J.Biol.Chem., 286, 2011
|
|
4I39
 
 | | Structures of ICT and PR1 intermediates from time-resolved laue crystallography collected at 14ID-B, APS | | 分子名称: | 4'-HYDROXYCINNAMIC ACID, Photoactive yellow protein | | 著者 | Jung, Y.O, Lee, J.H, Kim, J, Schmidt, M, Vukica, S, Moffat, K, Ihee, H. | | 登録日 | 2012-11-26 | | 公開日 | 2013-03-20 | | 最終更新日 | 2017-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Volume-conserving trans-cis isomerization pathways in photoactive yellow protein visualized by picosecond X-ray crystallography
NAT.CHEM., 5, 2013
|
|
6JKY
 
 | |
2ZOH
 
 | |
6J6H
 
 | | Cryo-EM structure of the yeast B*-a1 complex at an average resolution of 3.6 angstrom | | 分子名称: | ACT1 pre-mRNA, GUANOSINE-5'-TRIPHOSPHATE, INOSITOL HEXAKISPHOSPHATE, ... | | 著者 | Wan, R, Bai, R, Yan, C, Lei, J, Shi, Y. | | 登録日 | 2019-01-15 | | 公開日 | 2019-04-24 | | 最終更新日 | 2020-10-14 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Structures of the Catalytically Activated Yeast Spliceosome Reveal the Mechanism of Branching.
Cell, 177, 2019
|
|
2Z5U
 
 | |
4N7O
 
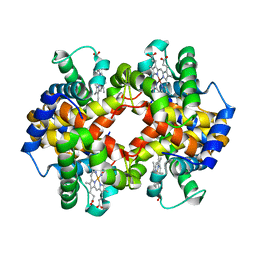 | | Capturing the haemoglobin allosteric transition in a single crystal form; Crystal structure of half-liganded human haemoglobin with phosphate at 2.5 A resolution. | | 分子名称: | Hemoglobin subunit alpha, Hemoglobin subunit beta, PROTOPORPHYRIN IX CONTAINING FE, ... | | 著者 | Sugiyama, K, Shibayama, N, Park, S.Y. | | 登録日 | 2013-10-16 | | 公開日 | 2014-04-02 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.502 Å) | | 主引用文献 | Capturing the hemoglobin allosteric transition in a single crystal form
J.Am.Chem.Soc., 136, 2014
|
|
4JRI
 
 | |
6J6Q
 
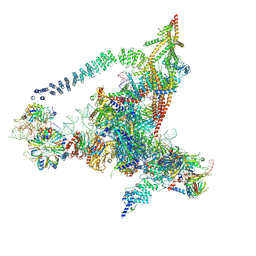 | | Cryo-EM structure of the yeast B*-b2 complex at an average resolution of 3.7 angstrom | | 分子名称: | GUANOSINE-5'-TRIPHOSPHATE, INOSITOL HEXAKISPHOSPHATE, MAGNESIUM ION, ... | | 著者 | Wan, R, Bai, R, Yan, C, Lei, J, Shi, Y. | | 登録日 | 2019-01-15 | | 公開日 | 2019-04-24 | | 最終更新日 | 2020-10-14 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Structures of the Catalytically Activated Yeast Spliceosome Reveal the Mechanism of Branching.
Cell, 177, 2019
|
|
6J6N
 
 | | Cryo-EM structure of the yeast B*-b1 complex at an average resolution of 3.86 angstrom | | 分子名称: | GUANOSINE-5'-TRIPHOSPHATE, INOSITOL HEXAKISPHOSPHATE, MAGNESIUM ION, ... | | 著者 | Wan, R, Bai, R, Yan, C, Lei, J, Shi, Y. | | 登録日 | 2019-01-15 | | 公開日 | 2019-04-24 | | 最終更新日 | 2020-10-14 | | 実験手法 | ELECTRON MICROSCOPY (3.86 Å) | | 主引用文献 | Structures of the Catalytically Activated Yeast Spliceosome Reveal the Mechanism of Branching.
Cell, 177, 2019
|
|
4IJG
 
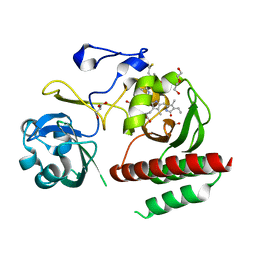 | | Crystal structure of monomeric bacteriophytochrome | | 分子名称: | 3-[2-[(Z)-[3-(2-carboxyethyl)-5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-4-methyl-pyrrol-1-ium -2-ylidene]methyl]-5-[(Z)-[(3E)-3-ethylidene-4-methyl-5-oxidanylidene-pyrrolidin-2-ylidene]methyl]-4-methyl-1H-pyrrol-3- yl]propanoic acid, Bacteriophytochrome, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Auldridge, M.E. | | 登録日 | 2012-12-21 | | 公開日 | 2013-12-25 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.701 Å) | | 主引用文献 | Origins of fluorescence in evolved bacteriophytochromes.
J.Biol.Chem., 289, 2014
|
|
6JON
 
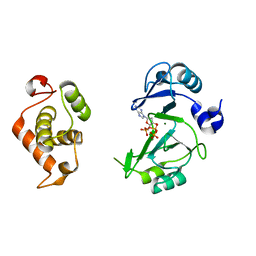 | | Crystal structures of phage NrS-1 N300-dNTPs-Mg2+ complex provide molecular mechanisms for substrate specificity | | 分子名称: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, MAGNESIUM ION, Primase | | 著者 | Guo, H.J, Li, M.J, Wu, H, Yu, F, He, J.H. | | 登録日 | 2019-03-22 | | 公開日 | 2019-06-26 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.34 Å) | | 主引用文献 | Crystal structures of phage NrS-1 N300-dNTPs-Mg2+complex provide molecular mechanisms for substrate specificity.
Biochem.Biophys.Res.Commun., 515, 2019
|
|
6JOP
 
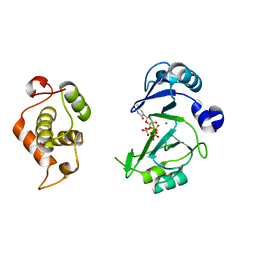 | | Crystal structures of phage NrS-1 N300-dNTPs-Mg2+ complex provide molecular mechanisms for substrate specificity | | 分子名称: | MAGNESIUM ION, Primase, THYMIDINE-5'-TRIPHOSPHATE | | 著者 | Guo, H.J, Li, M.J, Wu, H, Yu, F, He, J.H. | | 登録日 | 2019-03-22 | | 公開日 | 2019-06-26 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.353 Å) | | 主引用文献 | Crystal structures of phage NrS-1 N300-dNTPs-Mg2+complex provide molecular mechanisms for substrate specificity.
Biochem.Biophys.Res.Commun., 515, 2019
|
|
3CB5
 
 | | Crystal Structure of the S. pombe Peptidase Homology Domain of FACT complex subunit Spt16 (form A) | | 分子名称: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, FACT complex subunit spt16 | | 著者 | Stuwe, T, Hothorn, M, Lejeune, E, Bortfeld-Miller, M, Scheffzek, K, Ladurner, A.G. | | 登録日 | 2008-02-21 | | 公開日 | 2008-06-17 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | The FACT Spt16 "peptidase" domain is a histone H3-H4 binding module
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
7K5J
 
 | | Structure of an E1-E2-ubiquitin thioester mimetic | | 分子名称: | ADENOSINE MONOPHOSPHATE, Ubiquitin, Ubiquitin-activating enzyme E1 1, ... | | 著者 | Yuan, L, Lv, Z, Olsen, S.K. | | 登録日 | 2020-09-16 | | 公開日 | 2021-04-28 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (3.42 Å) | | 主引用文献 | Crystal structures of an E1-E2-ubiquitin thioester mimetic reveal molecular mechanisms of transthioesterification.
Nat Commun, 12, 2021
|
|
3C65
 
 | |
