2LW4
 
 | | Solution NMR Structure of Human Transcription Elongation Factor A protein 2, Central Domain, Northeast Structural Genomics Consortium (NESG) Target HR8682B | | 分子名称: | Transcription elongation factor A protein 2 | | 著者 | Eletsky, A, Wang, D, Kohan, E, Janjua, H, Xiao, R, Acton, T.B, Everett, J.K, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2012-07-20 | | 公開日 | 2012-09-19 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution NMR Structure of Human Transcription Elongation Factor A protein 2, Central Domain (CASP Target)
To be Published
|
|
1EFW
 
 | | Crystal structure of aspartyl-tRNA synthetase from Thermus thermophilus complexed to tRNAasp from Escherichia coli | | 分子名称: | ASPARTYL-TRNA, ASPARTYL-TRNA SYNTHETASE | | 著者 | Briand, C, Poterszman, A, Eiler, S, Webster, G, Thierry, J.-C, Moras, D. | | 登録日 | 2000-02-10 | | 公開日 | 2000-06-19 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | An intermediate step in the recognition of tRNA(Asp) by aspartyl-tRNA synthetase.
J.Mol.Biol., 299, 2000
|
|
2DJB
 
 | | Solution structure of the RING domain of the human Polycomb group RING finger protein 6 | | 分子名称: | Polycomb group RING finger protein 6, ZINC ION | | 著者 | Miyamoto, K, Kigawa, T, Sato, M, Koshiba, S, Inoue, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2006-03-31 | | 公開日 | 2007-03-13 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of the RING domain of the human Polycomb group RING finger protein 6
To be Published
|
|
1WPU
 
 | |
1WRO
 
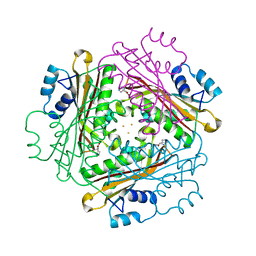 | | Metal Ion dependency of the antiterminator protein, HutP, for binding to the terminator region of hut mRNA- A structural basis | | 分子名称: | BARIUM ION, HISTIDINE, Hut operon positive regulatory protein | | 著者 | Kumarevel, T, Mizuno, H, Kumar, P.K.R. | | 登録日 | 2004-10-25 | | 公開日 | 2005-08-30 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | Characterization of the metal ion binding site in the anti-terminator protein, HutP, of Bacillus subtilis
Nucleic Acids Res., 33, 2005
|
|
1ECJ
 
 | |
7EZP
 
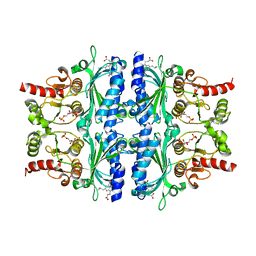 | | Indole-2-carboxylic acid derivatives as allosteric inhibitors of fructose-1,6-bisphosphatase | | 分子名称: | 1,6-di-O-phosphono-beta-D-fructofuranose, 3-(3-hydroxy-3-oxopropyl)-5-(2-methylpropyl)-7-nitro-1H-indole-2-carboxylic acid, Fructose-1,6-bisphosphatase 1 | | 著者 | Wang, X.Y, Zhou, J, Xu, B.L. | | 登録日 | 2021-06-01 | | 公開日 | 2022-06-01 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Discovery of Novel Indole Derivatives as Fructose-1,6-bisphosphatase Inhibitors and X-ray Cocrystal Structures Analysis.
Acs Med.Chem.Lett., 13, 2022
|
|
7EZF
 
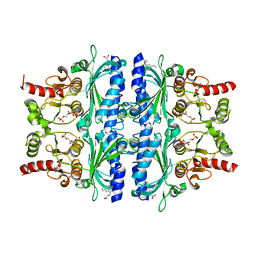 | | Indole-2-carboxylic acid derivatives as allosteric inhibitors of fructose-1,6-bisphosphatase | | 分子名称: | 1,6-di-O-phosphono-beta-D-fructofuranose, 7-chloranyl-5-ethyl-3-(3-hydroxy-3-oxopropyl)-1H-indole-2-carboxylic acid, Fructose-1,6-bisphosphatase 1 | | 著者 | Wang, X.Y, Zhou, J, Xu, B.L. | | 登録日 | 2021-06-01 | | 公開日 | 2022-06-01 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.76 Å) | | 主引用文献 | Discovery of Novel Indole Derivatives as Fructose-1,6-bisphosphatase Inhibitors and X-ray Cocrystal Structures Analysis.
Acs Med.Chem.Lett., 13, 2022
|
|
5DS3
 
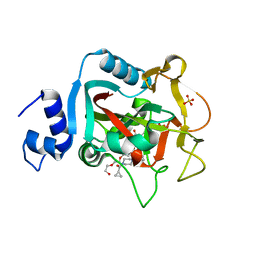 | | Crystal structure of constitutively active PARP-1 | | 分子名称: | 4-(3-{[4-(cyclopropylcarbonyl)piperazin-1-yl]carbonyl}-4-fluorobenzyl)phthalazin-1(2H)-one, PENTAETHYLENE GLYCOL, Poly [ADP-ribose] polymerase 1, ... | | 著者 | Langelier, M.F, Pascal, J.M. | | 登録日 | 2015-09-16 | | 公開日 | 2016-07-27 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | PARP-1 Activation Requires Local Unfolding of an Autoinhibitory Domain.
Mol.Cell, 60, 2015
|
|
7EZR
 
 | | Indole-2-carboxylic acid derivatives as allosteric inhibitors of fructose-1,6-bisphosphatase | | 分子名称: | 1,6-di-O-phosphono-beta-D-fructofuranose, 5-ethyl-7-nitro-3-[3-oxidanylidene-3-(thiophen-2-ylsulfonylamino)propyl]-1H-indole-2-carboxylic acid, Fructose-1,6-bisphosphatase 1 | | 著者 | Wang, X.Y, Zhou, J, Xu, B.L. | | 登録日 | 2021-06-01 | | 公開日 | 2022-06-01 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (3.27 Å) | | 主引用文献 | Discovery of Novel Indole Derivatives as Fructose-1,6-bisphosphatase Inhibitors and X-ray Cocrystal Structures Analysis.
Acs Med.Chem.Lett., 13, 2022
|
|
1WZ3
 
 | | The crystal structure of plant ATG12 | | 分子名称: | autophagy 12b | | 著者 | Suzuki, N.N, Yoshimoto, K, Fujioka, Y, Ohsumi, Y, Inagaki, F. | | 登録日 | 2005-02-22 | | 公開日 | 2005-06-21 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | The crystal structure of plant ATG12 and its biological implication in autophagy.
Autophagy, 1, 2005
|
|
3CDU
 
 | | Crystal structure of coxsackievirus B3 RNA-dependent RNA polymerase (3Dpol) in complex with a pyrophosphate | | 分子名称: | ACETATE ION, CHLORIDE ION, GLYCEROL, ... | | 著者 | Gruez, A, Selisko, B, Roberts, M, Bricogne, G, Bussetta, C, Canard, B. | | 登録日 | 2008-02-27 | | 公開日 | 2008-07-22 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | The crystal structure of coxsackievirus B3 RNA-dependent RNA polymerase in complex with its protein primer VPg confirms the existence of a second VPg binding site on Picornaviridae polymerases
J.Virol., 82, 2008
|
|
5DSY
 
 | | Crystal structure of constitutively active PARP-2 | | 分子名称: | 2-[4-[(2S,3S,4R,5R)-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]carbonylpiperazin-1-yl]-N-(1-oxidanylidene-2,3-dihydroisoindol-4-yl)ethanamide, Poly [ADP-ribose] polymerase 2 | | 著者 | Riccio, A.A, Pascal, J.M. | | 登録日 | 2015-09-17 | | 公開日 | 2016-07-27 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | PARP-1 Activation Requires Local Unfolding of an Autoinhibitory Domain.
Mol.Cell, 60, 2015
|
|
7EZW
 
 | |
7F07
 
 | | Autonomous VH domain that interacts with eIF4E at the Capped mRNA Binding site. | | 分子名称: | Eukaryotic translation initiation factor 4E, VH domain (VH-DiFCAP-01) | | 著者 | Brown, C.J, Frosi, Y, Ng, S, Lin, Y.C. | | 登録日 | 2021-06-03 | | 公開日 | 2022-06-08 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Development of a novel peptide aptamer that interacts with the eIF4E capped-mRNA binding site using peptide epitope linker evolution (PELE).
Rsc Chem Biol, 3, 2022
|
|
2C63
 
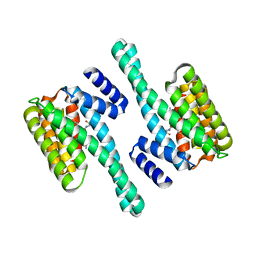 | | 14-3-3 Protein Eta (Human) Complexed to Peptide | | 分子名称: | 14-3-3 PROTEIN ETA, CONSENSUS PEPTIDE FOR 14-3-3 PROTEINS | | 著者 | Elkins, J.M, Yang, X, Smee, C.E.A, Johansson, C, Sundstrom, M, Edwards, A, Weigelt, J, Arrowsmith, C, Doyle, D.A. | | 登録日 | 2005-11-07 | | 公開日 | 2005-11-21 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | Structural Basis for Protein-Protein Interactions in the 14-3-3 Protein Family.
Proc.Natl.Acad.Sci.USA, 103, 2006
|
|
3CDW
 
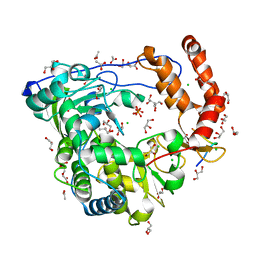 | | Crystal structure of coxsackievirus B3 RNA-dependent RNA polymerase (3Dpol) in complex with protein primer VPg and a pyrophosphate | | 分子名称: | ACETATE ION, CHLORIDE ION, GLYCEROL, ... | | 著者 | Gruez, A, Selisko, B, Roberts, M, Bricogne, G, Bussetta, C, Canard, B. | | 登録日 | 2008-02-27 | | 公開日 | 2008-07-22 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | The crystal structure of coxsackievirus B3 RNA-dependent RNA polymerase in complex with its protein primer VPg confirms the existence of a second VPg binding site on Picornaviridae polymerases
J.Virol., 82, 2008
|
|
7F3L
 
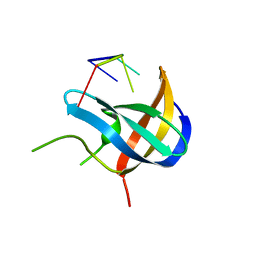 | |
1EJ0
 
 | | FTSJ RNA METHYLTRANSFERASE COMPLEXED WITH S-ADENOSYLMETHIONINE, MERCURY DERIVATIVE | | 分子名称: | FTSJ, MERCURY (II) ION, S-ADENOSYLMETHIONINE | | 著者 | Bugl, H, Fauman, E.B, Staker, B.L, Zheng, F, Kushner, S.R, Saper, M.A, Bardwell, J.C.A, Jakob, U. | | 登録日 | 2000-02-29 | | 公開日 | 2000-08-30 | | 最終更新日 | 2023-08-09 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | RNA methylation under heat shock control.
Mol.Cell, 6, 2000
|
|
7F3J
 
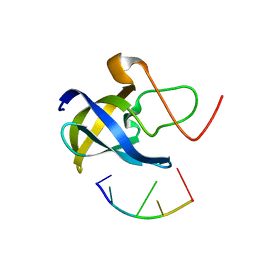 | |
1WLZ
 
 | | Crystal structure of DJBP fragment which was obtained by limited proteolysis | | 分子名称: | CAP-binding protein complex interacting protein 1 isoform a | | 著者 | Honbou, K, Suzuki, N, Horiuchi, M, Taira, T, Niki, T, Ariga, H, Inagaki, F. | | 登録日 | 2004-07-01 | | 公開日 | 2005-08-23 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Crystal Structure of DJBP Fragment which was obtained by Limited Proteolysis
To be Published
|
|
7F3I
 
 | |
7F3K
 
 | |
7FH9
 
 | |
3UEF
 
 | | Crystal structure of human Survivin bound to histone H3 (C2 space group). | | 分子名称: | 1,2-ETHANEDIOL, Baculoviral IAP repeat-containing protein 5, N-terminal fragment of histone H3, ... | | 著者 | Niedzialkowska, E, Porebski, P.J, Wang, F, Higgins, J.M, Stukenberg, P.T, Minor, W. | | 登録日 | 2011-10-30 | | 公開日 | 2012-03-07 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.45 Å) | | 主引用文献 | Molecular basis for phosphospecific recognition of histone H3 tails by Survivin paralogues at inner centromeres.
Mol.Biol.Cell, 23, 2012
|
|
