8III
 
 | |
8IIH
 
 | | H109C mutant of uracil DNA glycosylase X | | 分子名称: | IRON/SULFUR CLUSTER, Type-4 uracil-DNA glycosylase | | 著者 | Aroli, S. | | 登録日 | 2023-02-24 | | 公開日 | 2023-06-21 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (2.13 Å) | | 主引用文献 | Mutational and structural analyses of UdgX: insights into the active site pocket architecture and its evolution.
Nucleic Acids Res., 51, 2023
|
|
8IIG
 
 | |
8IIF
 
 | | H109A mutant of uracil DNA glycosylase X | | 分子名称: | GLYCEROL, IRON/SULFUR CLUSTER, Type-4 uracil-DNA glycosylase | | 著者 | Aroli, S. | | 登録日 | 2023-02-24 | | 公開日 | 2023-06-21 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Mutational and structural analyses of UdgX: insights into the active site pocket architecture and its evolution.
Nucleic Acids Res., 51, 2023
|
|
8IIE
 
 | |
8IIA
 
 | |
8II0
 
 | | FACTOR INHIBITING HIF-1 ALPHA in complex with (5-(3-(3-chlorophenyl)isoxazol-5-yl)-3-hydroxypicolinoyl)glycine | | 分子名称: | 2-[[5-[3-(3-chlorophenyl)-1,2-oxazol-5-yl]-3-oxidanyl-pyridin-2-yl]carbonylamino]ethanoic acid, GLYCEROL, Hypoxia-inducible factor 1-alpha inhibitor, ... | | 著者 | Nakashima, Y, Corner, T, Zhang, X, Schofield, C.J. | | 登録日 | 2023-02-24 | | 公開日 | 2024-02-28 | | 最終更新日 | 2024-07-10 | | 実験手法 | X-RAY DIFFRACTION (2.04 Å) | | 主引用文献 | A Small-Molecule Inhibitor of Factor Inhibiting HIF Binding to a Tyrosine-flip Pocket for the Treatment of Obesity.
Angew.Chem.Int.Ed.Engl., 2024
|
|
8IHY
 
 | | X-ray crystal structure of Q387E mutant of endo-1,4-beta glucanase from Eisenia fetida | | 分子名称: | CALCIUM ION, Endoglucanase, GLYCEROL, ... | | 著者 | Kuroki, C, Hirano, Y, Nakazawa, M, Sakamoto, T, Tamada, T, Ueda, M. | | 登録日 | 2023-02-24 | | 公開日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | A single mutation Asp43Arg was increased 2.5-fold the catalytic activity and maintained the stability of cold-adapted endo-1,4-beta glucanase (Ef-EG2) from Eisenia fetida.
Curr Res Biotechnol, 5, 2023
|
|
8IHX
 
 | | X-ray crystal structure of N372D mutant of endo-1,4-beta glucanase from Eisenia fetida | | 分子名称: | CALCIUM ION, Endoglucanase, GLYCEROL, ... | | 著者 | Kuroki, C, Hirano, Y, Nakazawa, M, Sakamoto, T, Tamada, T, Ueda, M. | | 登録日 | 2023-02-24 | | 公開日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | A single mutation Asp43Arg was increased 2.5-fold the catalytic activity and maintained the stability of cold-adapted endo-1,4-beta glucanase (Ef-EG2) from Eisenia fetida.
Curr Res Biotechnol, 5, 2023
|
|
8IHW
 
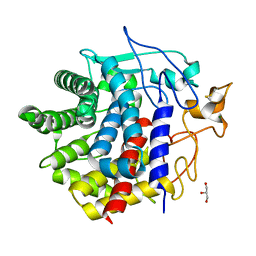 | | X-ray crystal structure of D43R mutant of endo-1,4-beta glucanase from Eisenia fetida | | 分子名称: | CALCIUM ION, Endoglucanase, GLYCEROL, ... | | 著者 | Kuroki, C, Hirano, Y, Nakazawa, M, Sakamoto, T, Tamada, T, Ueda, M. | | 登録日 | 2023-02-24 | | 公開日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | A single mutation Asp43Arg was increased 2.5-fold the catalytic activity and maintained the stability of cold-adapted endo-1,4-beta glucanase (Ef-EG2) from Eisenia fetida.
Curr Res Biotechnol, 5, 2023
|
|
8IHT
 
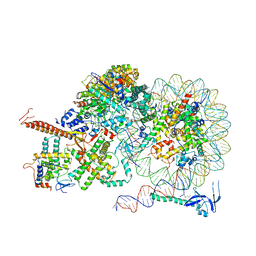 | | Rpd3S bound to the nucleosome | | 分子名称: | CALCIUM ION, Chromatin modification-related protein EAF3, DNA (164-MER), ... | | 著者 | Zhang, Y, Gang, C. | | 登録日 | 2023-02-23 | | 公開日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (3.72 Å) | | 主引用文献 | Structural basis for nucleosome binding and catalysis by the yeast Rpd3S/HDAC holoenzyme.
Cell Res., 33, 2023
|
|
8IHS
 
 | | Cryo-EM structure of ochratoxin A-detoxifying amidohydrolase ADH3 in complex with ochratoxin A | | 分子名称: | (2~{S})-2-[[(3~{R})-5-chloranyl-3-methyl-8-oxidanyl-1-oxidanylidene-3,4-dihydroisochromen-7-yl]carbonylamino]-3-phenyl-propanoic acid, Amidohydrolase family protein, ZINC ION | | 著者 | Dai, L.H, Niu, D, Huang, J.-W, Li, X, Shen, P.P, Li, H, Hu, Y.M, Yang, Y, Chen, C.-C, Guo, R.-T. | | 登録日 | 2023-02-23 | | 公開日 | 2023-08-30 | | 最終更新日 | 2023-11-15 | | 実験手法 | ELECTRON MICROSCOPY (2.5 Å) | | 主引用文献 | Cryo-EM structure and rational engineering of a superefficient ochratoxin A-detoxifying amidohydrolase.
J Hazard Mater, 458, 2023
|
|
8IHN
 
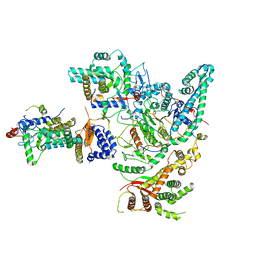 | | Cryo-EM structure of the Rpd3S core complex | | 分子名称: | CALCIUM ION, Chromatin modification-related protein EAF3, Histone H3, ... | | 著者 | Zhang, Y, Gang, C. | | 登録日 | 2023-02-23 | | 公開日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (3.37 Å) | | 主引用文献 | Structural basis for nucleosome binding and catalysis by the yeast Rpd3S/HDAC holoenzyme.
Cell Res., 33, 2023
|
|
8IHL
 
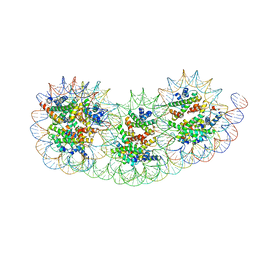 | | Overlapping tri-nucleosome | | 分子名称: | DNA (353-MER), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | 著者 | Nishimura, M, Fujii, T, Tanaka, H, Maehara, K, Nozawa, K, Takizawa, Y, Ohkawa, Y, Kurumizaka, H. | | 登録日 | 2023-02-23 | | 公開日 | 2024-01-17 | | 最終更新日 | 2024-01-24 | | 実験手法 | ELECTRON MICROSCOPY (7.64 Å) | | 主引用文献 | Genome-wide mapping and cryo-EM structural analyses of the overlapping tri-nucleosome composed of hexasome-hexasome-octasome moieties.
Commun Biol, 7, 2024
|
|
8IHK
 
 | | Cryo-EM structure of HCA3-Gi complex with acifran (local) | | 分子名称: | (5~{S})-5-methyl-4-oxidanylidene-5-phenyl-furan-2-carboxylic acid, Soluble cytochrome b562,Hydroxycarboxylic acid receptor 3 | | 著者 | Suzuki, S, Nishikawa, K, Suzuki, H, Fujiyoshi, Y. | | 登録日 | 2023-02-22 | | 公開日 | 2023-08-30 | | 最終更新日 | 2023-12-06 | | 実験手法 | ELECTRON MICROSCOPY (3.21 Å) | | 主引用文献 | Structural basis of hydroxycarboxylic acid receptor signaling mechanisms through ligand binding.
Nat Commun, 14, 2023
|
|
8IHJ
 
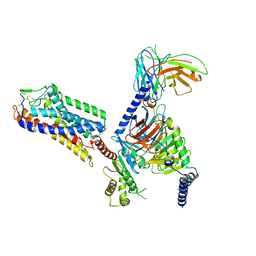 | | Cryo-EM structure of HCA3-Gi complex with acifran | | 分子名称: | (5~{S})-5-methyl-4-oxidanylidene-5-phenyl-furan-2-carboxylic acid, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | 著者 | Suzuki, S, Nishikawa, K, Suzuki, H, Fujiyoshi, Y. | | 登録日 | 2023-02-22 | | 公開日 | 2023-08-30 | | 最終更新日 | 2023-12-06 | | 実験手法 | ELECTRON MICROSCOPY (3.07 Å) | | 主引用文献 | Structural basis of hydroxycarboxylic acid receptor signaling mechanisms through ligand binding.
Nat Commun, 14, 2023
|
|
8IHI
 
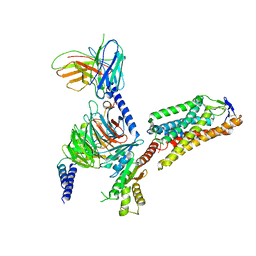 | | Cryo-EM structure of HCA2-Gi complex with acifran | | 分子名称: | (5~{S})-5-methyl-4-oxidanylidene-5-phenyl-furan-2-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | 著者 | Suzuki, S, Nishikawa, K, Suzuki, H, Fujiyoshi, Y. | | 登録日 | 2023-02-22 | | 公開日 | 2023-08-30 | | 最終更新日 | 2023-12-06 | | 実験手法 | ELECTRON MICROSCOPY (3.11 Å) | | 主引用文献 | Structural basis of hydroxycarboxylic acid receptor signaling mechanisms through ligand binding.
Nat Commun, 14, 2023
|
|
8IHH
 
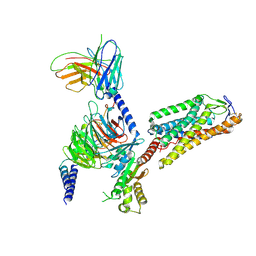 | | Cryo-EM structure of HCA2-Gi complex with LUF6283 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 5-butyl-1~{H}-pyrazole-3-carboxylic acid, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | 著者 | Suzuki, S, Nishikawa, K, Suzuki, H, Fujiyoshi, Y. | | 登録日 | 2023-02-22 | | 公開日 | 2023-08-30 | | 最終更新日 | 2023-12-06 | | 実験手法 | ELECTRON MICROSCOPY (3.06 Å) | | 主引用文献 | Structural basis of hydroxycarboxylic acid receptor signaling mechanisms through ligand binding.
Nat Commun, 14, 2023
|
|
8IHG
 
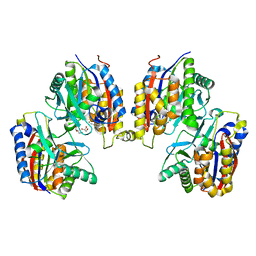 | |
8IHF
 
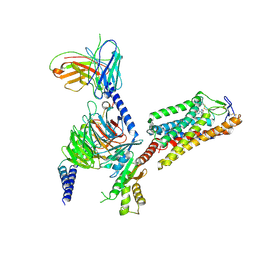 | | Cryo-EM structure of HCA2-Gi complex with MK6892 | | 分子名称: | 2-[[2,2-dimethyl-3-[3-(5-oxidanylpyridin-2-yl)-1,2,4-oxadiazol-5-yl]propanoyl]amino]cyclohexene-1-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | 著者 | Suzuki, S, Nishikawa, K, Suzuki, H, Fujiyoshi, Y. | | 登録日 | 2023-02-22 | | 公開日 | 2023-08-30 | | 最終更新日 | 2023-12-06 | | 実験手法 | ELECTRON MICROSCOPY (2.97 Å) | | 主引用文献 | Structural basis of hydroxycarboxylic acid receptor signaling mechanisms through ligand binding.
Nat Commun, 14, 2023
|
|
8IHB
 
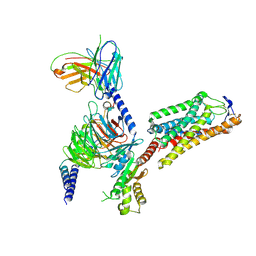 | | Cryo-EM structure of HCA2-Gi complex with GSK256073 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 8-chloranyl-3-pentyl-7H-purine-2,6-dione, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | 著者 | Suzuki, S, Nishikawa, K, Suzuki, H, Fujiyoshi, Y. | | 登録日 | 2023-02-22 | | 公開日 | 2023-09-13 | | 最終更新日 | 2023-12-06 | | 実験手法 | ELECTRON MICROSCOPY (2.85 Å) | | 主引用文献 | Structural basis of hydroxycarboxylic acid receptor signaling mechanisms through ligand binding.
Nat Commun, 14, 2023
|
|
8IH8
 
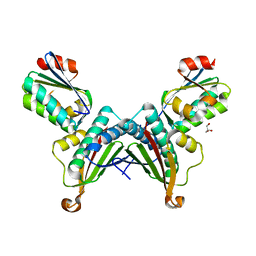 | |
8IH6
 
 | |
8IH5
 
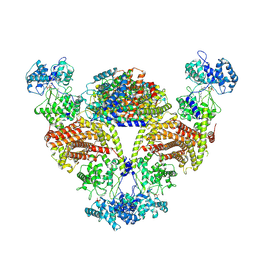 | |
8IH1
 
 | |
