1Y6J
 
 | | L-Lactate Dehydrogenase from Clostridium Thermocellum Cth-1135 | | 分子名称: | L-lactate dehydrogenase | | 著者 | Chen, L, Yang, H, Kataeva, I, Chen, L.R, Tempel, W, Lee, D, Habel, J, Zhou, W, Lin, D, Ljungdahl, L, Liu, Z.-J, Rose, J, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG) | | 登録日 | 2004-12-06 | | 公開日 | 2004-12-14 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (3.01 Å) | | 主引用文献 | L-Lactate Dehydrogenase from Clostridium Thermocellum Cth-1135
To be Published
|
|
2BAA
 
 | | THE REFINED CRYSTAL STRUCTURE OF AN ENDOCHITINASE FROM HORDEUM VULGARE L. SEEDS TO 1.8 ANGSTROMS RESOLUTION | | 分子名称: | ENDOCHITINASE (26 KD) | | 著者 | Hart, P.J, Pfluger, H.D, Monzingo, A.F, Ready, M.P, Ernst, S.R, Hollis, T, Robertus, J.D. | | 登録日 | 1995-01-26 | | 公開日 | 1996-01-15 | | 最終更新日 | 2024-06-05 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | The refined crystal structure of an endochitinase from Hordeum vulgare L. seeds at 1.8 A resolution.
J.Mol.Biol., 248, 1995
|
|
4CEI
 
 | | Crystal structure of ADPNP-bound AddAB with a forked DNA substrate | | 分子名称: | ATP-DEPENDENT HELICASE/DEOXYRIBONUCLEASE SUBUNIT B, ATP-DEPENDENT HELICASE/NUCLEASE SUBUNIT A, DNA, ... | | 著者 | Krajewski, W.W, Wilkinson, M, Fu, X, Cronin, N.B, Wigley, D. | | 登録日 | 2013-11-11 | | 公開日 | 2014-03-12 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Structural Basis for Translocation by Addab Helicase-Nuclease and its Arrest at Chi Sites.
Nature, 508, 2014
|
|
3ZOA
 
 | | The structure of Trehalose Synthase (TreS) of Mycobacterium smegmatis in complex with acarbose | | 分子名称: | 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, CALCIUM ION, CHLORIDE ION, ... | | 著者 | Caner, S, Nguyen, N, Aguda, A, Zhang, R, Pan, Y.T, Withers, S.G, Brayer, G.D. | | 登録日 | 2013-02-21 | | 公開日 | 2013-07-17 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | The Structure of the Mycobacterium Smegmatis Trehalose Synthase Reveals an Unusual Active Site Configuration and Acarbose-Binding Mode.
Glycobiology, 23, 2013
|
|
2XN0
 
 | | Structure of alpha-galactosidase from Lactobacillus acidophilus NCFM, PtCl4 derivative | | 分子名称: | ALPHA-GALACTOSIDASE, GLYCEROL, PLATINUM (II) ION | | 著者 | Fredslund, F, Abou Hachem, M, Larsen, R.J, Sorensen, P.G, Lo Leggio, L, Svensson, B. | | 登録日 | 2010-07-30 | | 公開日 | 2011-08-10 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Crystal Structure of Alpha-Galactosidase from Lactobacillus Acidophilus Ncfm: Insight Into Tetramer Formation and Substrate Binding.
J.Mol.Biol., 412, 2011
|
|
4CFS
 
 | | CRYSTAL STRUCTURE OF THE COFACTOR-DEVOID 1-H-3-HYDROXY-4- OXOQUINALDINE 2,4-DIOXYGENASE (HOD) CATALYTICALLY INACTIVE H251A VARIANT COMPLEXED WITH ITS NATURAL SUBSTRATE 1-H-3-HYDROXY-4- OXOQUINALDINE | | 分子名称: | 1-H-3-HYDROXY-4-OXOQUINALDINE 2,4-DIOXYGENASE, 3-HYDROXY-2-METHYLQUINOLIN-4(1H)-ONE, D(-)-TARTARIC ACID, ... | | 著者 | Bui, S, Steiner, R.A. | | 登録日 | 2013-11-19 | | 公開日 | 2013-12-04 | | 最終更新日 | 2014-04-02 | | 実験手法 | X-RAY DIFFRACTION (1.94 Å) | | 主引用文献 | Origin of the Proton-Transfer Step in the Cofactor-Free 1-H-3-Hydroxy-4-Oxoquinaldine 2,4- Dioxygenase: Effect of the Basicity of an Active Site His Residue.
J.Biol.Chem., 289, 2014
|
|
3ZF2
 
 | | Phage dUTPases control transfer of virulence genes by a proto- oncogenic G protein-like mechanism. (Staphylococcus bacteriophage 80alpha dUTPase). | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DUTPASE, NICKEL (II) ION | | 著者 | Tormo-Mas, M.A, Donderis, J, Garcia-Caballer, M, Alt, A, Mir-Sanchis, I, Marina, A, Penades, J.R. | | 登録日 | 2012-12-10 | | 公開日 | 2013-01-30 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Phage Dutpases Control Transfer of Virulence Genes by a Proto-Oncogenic G Protein-Like Mechanism.
Mol.Cell, 49, 2013
|
|
3ZJK
 
 | | crystal structure of Ttb-gly F401S mutant | | 分子名称: | BETA GLYCOSIDASE, CHLORIDE ION, GLYCEROL | | 著者 | Teze, D, Tran, V, Tellier, C, Dion, M, Leroux, C, Roncza, J, Czjzek, M. | | 登録日 | 2013-01-18 | | 公開日 | 2013-02-13 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Semi-Rational Approach for Converting a Gh1 Beta-Glycosidase Into a Beta-Transglycosidase.
Protein Eng.Des.Sel., 27, 2014
|
|
4BO2
 
 | | Crystal structure of 3-oxoacyl-(acyl-carrier-protein) reductase (FabG) from Pseudomonas aeruginosa in complex with 1-(1-ethylbenzimidazol-2- yl)-3-(2-methoxyphenyl)urea at 1.9A resolution | | 分子名称: | 1-(1-ethylbenzimidazol-2-yl)-3-(2-methoxyphenyl)urea, 3-OXOACYL-[ACYL-CARRIER-PROTEIN] REDUCTASE FABG | | 著者 | Cukier, C.D, Schnell, R, Lindqvist, Y, Schneider, G. | | 登録日 | 2013-05-18 | | 公開日 | 2013-09-18 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Discovery of an Allosteric Inhibitor Binding Site in 3-Oxo-Acyl-Acp Reductase from Pseudomonas Aeruginosa
Acs Chem.Biol., 8, 2013
|
|
3ZN2
 
 | | protein engineering of halohydrin dehalogenase | | 分子名称: | 1-METHOXY-2-[2-(2-METHOXY-ETHOXY]-ETHANE, ACETATE ION, HALOHYDRIN DEHALOGENASE, ... | | 著者 | Schallmey, M, Jekel, P, Tang, L, Majeric-Elenkov, M, Hoeffken, H.W, Hauer, B, Janssen, D.B. | | 登録日 | 2013-02-13 | | 公開日 | 2014-03-05 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | A Single Point Mutation Enhances Hydroxynitrile Synthesis by Halohydrin Dehalogenase.
Enzyme.Microb.Technol., 70, 2015
|
|
2Y6K
 
 | | Xylotetraose bound to X-2 engineered mutated CBM4-2 Carbohydrate Binding Module from a Thermostable Rhodothermus marinus Xylanase | | 分子名称: | CALCIUM ION, CITRIC ACID, XYLANASE, ... | | 著者 | von Schantz, L, Hakansson, M, Logan, D.T, Walse, B, Osterlin, J, Nordberg-Karlsson, E, Ohlin, M. | | 登録日 | 2011-01-24 | | 公開日 | 2012-03-07 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.36 Å) | | 主引用文献 | Structural basis for carbohydrate-binding specificity--a comparative assessment of two engineered carbohydrate-binding modules.
Glycobiology, 22, 2012
|
|
4C28
 
 | | Crystal structure of Trypanosoma cruzi CYP51 bound to the inhibitor (R)-N-(3-(1H-indol-3-yl)-1-oxo-1-(pyridin-4-ylamino)propan-2-yl)-4-(4-(4-chlorophenyl)piperazin-1-yl)-2-fluorobenzamide. | | 分子名称: | (R)-N-(3-(1H-indol-3-yl)-1-oxo-1-(pyridin-4-ylamino)propan-2-yl)-4-(4-(4-chlorophenyl)piperazin-1-yl)-2-fluorobenzamide, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | 著者 | Vieira, D.F, Calvet, C.M, Choi, J.Y, Cameron, M.D, Gut, J, Kellar, D, Siqueira-Neto, J.L, McKerrow, J.H, Roush, W.R, Podust, L.M. | | 登録日 | 2013-08-16 | | 公開日 | 2014-09-03 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.03 Å) | | 主引用文献 | Binding Mode and Potency of N-Indolyl-Oxopyridinyl-4-Amino-Propanyl-Based Inhibitors Targeting Trypanosoma Cruzi Cyp51
J.Med.Chem., 57, 2014
|
|
1YIF
 
 | | CRYSTAL STRUCTURE OF beta-1,4-xylosidase FROM BACILLUS SUBTILIS, NEW YORK STRUCTURAL GENOMICS CONSORTIUM | | 分子名称: | BETA-1,4-XYLOSIDASE | | 著者 | Patskovsky, Y, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | 登録日 | 2005-01-11 | | 公開日 | 2005-01-18 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | CRYSTAL STRUCTURE OF beta-1,4-xylosidase FROM BACILLUS SUBTILIS
To be Published
|
|
1OGS
 
 | | human acid-beta-glucosidase | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glucosylceramidase, ... | | 著者 | Dvir, H, Harel, M, McCarthy, A.A, Toker, L, Silman, I, Futerman, A.H, Sussman, J.L. | | 登録日 | 2003-05-13 | | 公開日 | 2003-07-03 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | X-Ray Structure of Human Acid-Beta-Glucosidase, the Defective Enzyme in Gaucher Disease
Embo Rep., 4, 2003
|
|
4CEJ
 
 | | Crystal structure of AddAB-DNA-ADPNP complex at 3 Angstrom resolution | | 分子名称: | ATP-DEPENDENT HELICASE/DEOXYRIBONUCLEASE SUBUNIT B, ATP-DEPENDENT HELICASE/NUCLEASE SUBUNIT A, DNA, ... | | 著者 | Krajewski, W.W, Wilkinson, M, Fu, X, Cronin, N.B, Wigley, D. | | 登録日 | 2013-11-11 | | 公開日 | 2014-03-12 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Structural Basis for Translocation by Addab Helicase-Nuclease and its Arrest at Chi Sites.
Nature, 508, 2014
|
|
4GIC
 
 | | Crystal Structure Of a Putative Histidinol dehydrogenase (Target PSI-014034) from Methylococcus capsulatus | | 分子名称: | Histidinol dehydrogenase, SULFATE ION | | 著者 | Kumar, P.R, Ahmed, M, Banu, N, Bhosle, R, Bonanno, J, Chamala, S, Chowdhury, S, Gizzi, A, Glen, S, Hammonds, J, Hillerich, B, Love, J.D, Seidel, R, Stead, M, Toro, R, Washington, E, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | 登録日 | 2012-08-08 | | 公開日 | 2012-08-22 | | 最終更新日 | 2017-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.052 Å) | | 主引用文献 | Crystal structure of a putative Histidinol dehydrogenase from Methylococcus capsulatus
to be published
|
|
1OJK
 
 | | Anatomy of glycosynthesis: Structure and kinetics of the Humicola insolens Cel7BE197A and E197S glycosynthase mutants | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ENDOGLUCANASE I, GLYCEROL, ... | | 著者 | Ducros, V.M.-A, Tarling, C.A, Zechel, D.L, Brzozowski, A.M, Frandsen, T.P, Von Ossowski, I, Schulein, M, Withers, S.G, Davies, G.J. | | 登録日 | 2003-07-10 | | 公開日 | 2004-01-07 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Anatomy of Glycosynthesis: Structure and Kinetics of the Humicola Insolens Cel7B E197A and E197S Glycosynthase Mutants
Chem.Biol., 10, 2003
|
|
4G09
 
 | |
3ZV4
 
 | | CRYSTAL STRUCTURE OF CIS-BIPHENYL-2,3-DIHYDRODIOL-2,3-DEHYDROGENASE (BPHB) FROM PANDORAEA PNOMENUSA STRAIN B-356 IN APO FORM AT 1.8 ANGSTROM | | 分子名称: | CIS-2,3-DIHYDROBIPHENYL-2,3-DIOL DEHYDROGENASE | | 著者 | Dhindwal, S, Patil, D.N, Kumar, P. | | 登録日 | 2011-07-23 | | 公開日 | 2011-08-31 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Biochemical Studies and Ligand-Bound Structures of Biphenyl Dehydrogenase from Pandoraea Pnomenusa Strain B-356 Reveal a Basis for Broad Specificity of the Enzyme.
J.Biol.Chem., 286, 2011
|
|
4BTY
 
 | | Crystal structure of human vascular adhesion protein-1 in complex with pyridazinone inhibitors | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 5-[4-(4-methylpiperazin-1-yl)phenylamino]-2-(4-chlorophenyl)-6-(1H-1,2,4-triazol-5-yl)-3(2H)-pyridazinone, ... | | 著者 | Bligt-Linden, E, Pihlavisto, M, Szatmari, I, Otwinowski, Z, Smith, D.J, Lazar, L, Fulop, F, Salminen, T.A. | | 登録日 | 2013-06-19 | | 公開日 | 2013-12-18 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Novel Pyridazinone Inhibitors for Vascular Adhesion Protein- 1 (Vap-1): Old Target - New Inhibition Mode.
J.Med.Chem., 56, 2013
|
|
1YKI
 
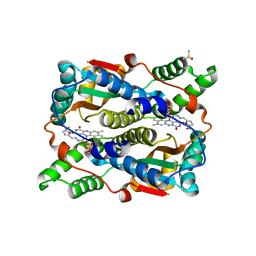 | | The structure of E. coli nitroreductase bound with the antibiotic nitrofurazone | | 分子名称: | CITRIC ACID, DIMETHYL SULFOXIDE, FLAVIN MONONUCLEOTIDE, ... | | 著者 | Race, P.R, Lovering, A.L, Green, R.M, Ossor, A, White, S.A, Searle, P.F, Wrighton, C.J, Hyde, E.I. | | 登録日 | 2005-01-18 | | 公開日 | 2005-02-08 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structural and mechanistic studies of Escherichia coli nitroreductase with the antibiotic nitrofurazone. Reversed binding orientations in different redox states of the enzyme.
J.Biol.Chem., 280, 2005
|
|
5QJ1
 
 | |
1YBA
 
 | |
1YKJ
 
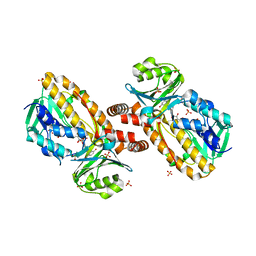 | | A45G p-hydroxybenzoate hydroxylase with p-hydroxybenzoate bound | | 分子名称: | FLAVIN-ADENINE DINUCLEOTIDE, P-HYDROXYBENZOIC ACID, P-hydroxybenzoate hydroxylase, ... | | 著者 | Cole, L.J, Gatti, D.L, Entsch, B, Ballou, D.P. | | 登録日 | 2005-01-18 | | 公開日 | 2005-07-26 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Removal of a methyl group causes global changes in p-hydroxybenzoate hydroxylase.
Biochemistry, 44, 2005
|
|
1OJI
 
 | | Anatomy of glycosynthesis: Structure and kinetics of the Humicola insolens Cel7B E197A and E197S glycosynthase mutants | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ENDOGLUCANASE I, GLYCEROL | | 著者 | Ducros, V.M.-A, Tarling, C.A, Zechel, D.L, Brzozowski, A.M, Frandsen, T.P, Von Ossowski, I, Schulein, M, Withers, S.G, Davies, G.J. | | 登録日 | 2003-07-10 | | 公開日 | 2004-01-07 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | Anatomy of Glycosynthesis: Structure and Kinetics of the Humicola Insolens Cel7B E197A and E197S Glycosynthase Mutants
Chem.Biol., 10, 2003
|
|
