6THE
 
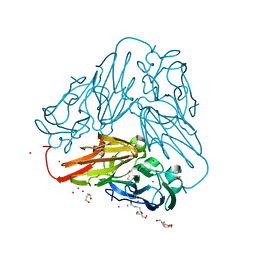 | | Crystal structure of core domain of four-domain heme-cupredoxin-Cu nitrite reductase from Bradyrhizobium sp. ORS 375 | | 分子名称: | CHLORIDE ION, COPPER (II) ION, Copper-containing nitrite reductase, ... | | 著者 | Sasaki, D, Watanabe, T.F, Eady, R.R, Garratt, R.C, Antonyuk, S.V, Hasnain, S.S. | | 登録日 | 2019-11-20 | | 公開日 | 2020-04-22 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.87 Å) | | 主引用文献 | Reverse protein engineering of a novel 4-domain copper nitrite reductase reveals functional regulation by protein-protein interaction.
Febs J., 288, 2021
|
|
6S0W
 
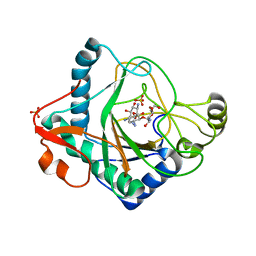 | | The crystal structure of kanamycin B dioxygenase (KanJ) from Streptomyces kanamyceticus in complex with nickel and kanamycin B sulfate | | 分子名称: | (1R,2S,3S,4R,6S)-4,6-DIAMINO-3-[(3-AMINO-3-DEOXY-ALPHA-D-GLUCOPYRANOSYL)OXY]-2-HYDROXYCYCLOHEXYL 2,6-DIAMINO-2,6-DIDEOXY-ALPHA-D-GLUCOPYRANOSIDE, DI(HYDROXYETHYL)ETHER, Kanamycin B dioxygenase, ... | | 著者 | Mrugala, B, Niedzialkowska, E, Minor, W, Borowski, T. | | 登録日 | 2019-06-18 | | 公開日 | 2020-07-08 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.36 Å) | | 主引用文献 | A study on the structure, mechanism, and biochemistry of kanamycin B dioxygenase (KanJ)-an enzyme with a broad range of substrates.
Febs J., 288, 2021
|
|
6THF
 
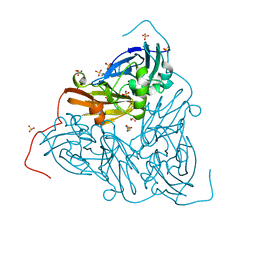 | | Crystal structure of two-domain Cu nitrite reductase from Bradyrhizobium sp. ORS 375 | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, COPPER (II) ION, Copper-containing nitrite reductase, ... | | 著者 | Sasaki, D, Watanabe, T.F, Eady, R.R, Garratt, R.C, Antonyuk, S.V, Hasnain, S.S. | | 登録日 | 2019-11-20 | | 公開日 | 2020-04-22 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.47 Å) | | 主引用文献 | Reverse protein engineering of a novel 4-domain copper nitrite reductase reveals functional regulation by protein-protein interaction.
Febs J., 288, 2021
|
|
6R82
 
 | |
6S0R
 
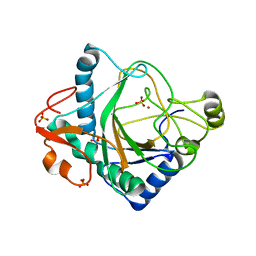 | | The crystal structure of kanamycin B dioxygenase (KanJ) from Streptomyces kanamyceticus complex with nickel, sulfate and chloride | | 分子名称: | CHLORIDE ION, Kanamycin B dioxygenase, NICKEL (II) ION, ... | | 著者 | Mrugala, B, Porebski, P.J, Niedzialkowska, E, Cymborowski, M.T, Minor, W, Borowski, T. | | 登録日 | 2019-06-18 | | 公開日 | 2020-07-08 | | 最終更新日 | 2024-06-19 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | A study on the structure, mechanism, and biochemistry of kanamycin B dioxygenase (KanJ)-an enzyme with a broad range of substrates.
Febs J., 288, 2021
|
|
6S1V
 
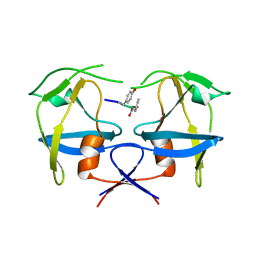 | | Crystal structure of dimeric M-PMV protease D26N mutant in complex with inhibitor | | 分子名称: | Gag-Pro-Pol polyprotein, PRO-0A1-VAL-PSA-ALA-MET-THR | | 著者 | Wosicki, S, Gilski, M, Jaskolski, M, Zabranska, H, Pichova, I. | | 登録日 | 2019-06-19 | | 公開日 | 2019-10-16 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.64 Å) | | 主引用文献 | Comparison of a retroviral protease in monomeric and dimeric states.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
6S0U
 
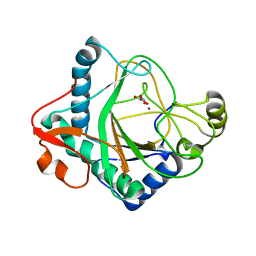 | | The crystal structure of kanamycin B dioxygenase (KanJ) from Streptomyces kanamyceticus in complex with nickel and 2-oxoglutarate | | 分子名称: | 2-OXOGLUTARIC ACID, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Mrugala, B, Porebski, P.J, Niedzialkowska, E, Minor, W, Borowski, T. | | 登録日 | 2019-06-18 | | 公開日 | 2020-07-08 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | A study on the structure, mechanism, and biochemistry of kanamycin B dioxygenase (KanJ)-an enzyme with a broad range of substrates.
Febs J., 288, 2021
|
|
6S1U
 
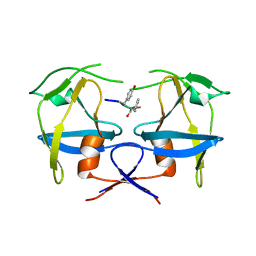 | | Crystal structure of dimeric M-PMV protease C7A/D26N/C106A mutant in complex with inhibitor | | 分子名称: | Gag-Pro-Pol polyprotein, PRO-0A1-VAL-PSA-ALA-MET-THR | | 著者 | Wosicki, S, Gilski, M, Jaskolski, M, Zabranska, H, Pichova, I. | | 登録日 | 2019-06-19 | | 公開日 | 2019-10-16 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Comparison of a retroviral protease in monomeric and dimeric states.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
6S1W
 
 | | Crystal structure of dimeric M-PMV protease D26N mutant | | 分子名称: | Gag-Pro-Pol polyprotein | | 著者 | Wosicki, S, Gilski, M, Jaskolski, M, Zabranska, H, Pichova, I. | | 登録日 | 2019-06-19 | | 公開日 | 2019-10-16 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.98 Å) | | 主引用文献 | Comparison of a retroviral protease in monomeric and dimeric states.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
6S0T
 
 | | The crystal structure of kanamycin B dioxygenase (KanJ) from Streptomyces kanamyceticus in complex with nickel, sulfate, soaked with iodide | | 分子名称: | IODIDE ION, Kanamycin B dioxygenase, NICKEL (II) ION, ... | | 著者 | Mrugala, B, Porebski, P.J, Niedzialkowska, E, Cymborowski, M.T, Minor, W, Borowski, T. | | 登録日 | 2019-06-18 | | 公開日 | 2020-07-08 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | A study on the structure, mechanism, and biochemistry of kanamycin B dioxygenase (KanJ)-an enzyme with a broad range of substrates.
Febs J., 288, 2021
|
|
6SGF
 
 | | Molecular insight into a new low affinity xylan binding module CBM86, from the xylanolytic gut symbiont Roseburia intestinalis. | | 分子名称: | Beta-xylanase, CADMIUM ION, CHLORIDE ION, ... | | 著者 | Ejby, M, Abou Hachem, M, Leth, M.L, Guskov, A, Slotboom, D. | | 登録日 | 2019-08-04 | | 公開日 | 2019-11-20 | | 最終更新日 | 2024-05-15 | | 実験手法 | X-RAY DIFFRACTION (1.756 Å) | | 主引用文献 | Molecular insight into a new low-affinity xylan binding module from the xylanolytic gut symbiont Roseburia intestinalis.
Febs J., 287, 2020
|
|
5CFY
 
 | |
5DT1
 
 | |
7F3Y
 
 | | Wild-type Plasmodium falciparum dihydrofolate reductase-thymidylate synthase (PfDHFR-TS) complexed with methotrexate (MTX), NADPH and dUMP | | 分子名称: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, Bifunctional dihydrofolate reductase-thymidylate synthase, GLYCEROL, ... | | 著者 | Vanichtanankul, J, Tanramluk, D, Yuvaniyama, J, Yuthavong, Y. | | 登録日 | 2021-06-17 | | 公開日 | 2021-09-22 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.252 Å) | | 主引用文献 | MANORAA: A machine learning platform to guide protein-ligand design by anchors and influential distances.
Structure, 30, 2022
|
|
7F3Z
 
 | | Double mutant Plasmodium falciparum dihydrofolate reductase-thymidylate synthase (PfDHFR-TS-K1, C59R+S108N) complexed with Trimethoprim (TOP), NADPH and dUMP | | 分子名称: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, Bifunctional dihydrofolate reductase-thymidylate synthase, GLYCEROL, ... | | 著者 | Vanichtanankul, J, Tanramluk, D, Chitnumsub, P, Yuvaniyama, J, Yuthavong, Y. | | 登録日 | 2021-06-17 | | 公開日 | 2021-09-22 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | MANORAA: A machine learning platform to guide protein-ligand design by anchors and influential distances.
Structure, 30, 2022
|
|
7B8E
 
 | | Torpedo californica acetylcholinesterase complexed with Ca+2 | | 分子名称: | 1,2-ETHANEDIOL, 2-[2-(2-ethoxyethoxy)ethoxy]ethanol, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Silman, I, Shnyrov, V.L, Ashani, Y, Roth, E, Nicolas, A, Sussman, J.L. | | 登録日 | 2020-12-12 | | 公開日 | 2021-03-17 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.23 Å) | | 主引用文献 | Torpedo californica acetylcholinesterase is stabilized by binding of a divalent metal ion to a novel and versatile 4D motif.
Protein Sci., 30, 2021
|
|
7DFS
 
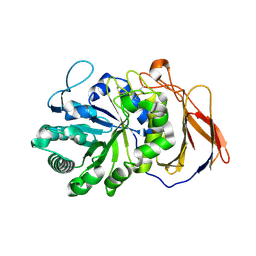 | | Crystal structure of a novel 4-O-alpha-L-rhamnosyl-beta-D-glucuronidase from Fusarium oxysporum 12S - Rha-GlcA complex | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-O-alpha-L-rhamnosyl-beta-D-glucuronidase, alpha-D-mannopyranose, ... | | 著者 | Kondo, T, Arakawa, T, Fushinobu, S, Sakamoto, T. | | 登録日 | 2020-11-09 | | 公開日 | 2021-03-17 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.49 Å) | | 主引用文献 | Biochemical and structural characterization of a novel 4-O-alpha-l-rhamnosyl-beta-d-glucuronidase from Fusarium oxysporum.
Febs J., 288, 2021
|
|
7DFQ
 
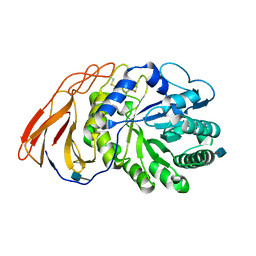 | | Crystal Structure of a novel 4-O-alpha-L-rhamnosyl-beta-D-glucuronidase from Fusarium oxysporum 12S, ligand-free form | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-O-alpha-L-rhamnosyl-beta-D-glucuronidase | | 著者 | Kondo, T, Arakawa, T, Fushinobu, S, Sakamoto, T. | | 登録日 | 2020-11-09 | | 公開日 | 2021-03-17 | | 最終更新日 | 2021-08-25 | | 実験手法 | X-RAY DIFFRACTION (1.51 Å) | | 主引用文献 | Biochemical and structural characterization of a novel 4-O-alpha-l-rhamnosyl-beta-d-glucuronidase from Fusarium oxysporum.
Febs J., 288, 2021
|
|
6FJ2
 
 | |
6FJ9
 
 | |
3J2W
 
 | |
3J4R
 
 | |
3J2Y
 
 | |
6LCE
 
 | |
6LCF
 
 | |
