8OFB
 
 | | Crystal Structure of T. maritima reverse gyrase with a minimal latch, hexagonal form | | 分子名称: | CHLORIDE ION, HEXAETHYLENE GLYCOL, Reverse gyrase, ... | | 著者 | Klostermeier, D, Rasche, R, Mhaindarkar, V, Kummel, D, Rudolph, M.G. | | 登録日 | 2023-03-15 | | 公開日 | 2023-04-26 | | 最終更新日 | 2024-06-19 | | 実験手法 | X-RAY DIFFRACTION (2.39 Å) | | 主引用文献 | Structure of reverse gyrase with a minimal latch that supports ATP-dependent positive supercoiling without specific interactions with the topoisomerase domain.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
1YD0
 
 | | Crystal structure of the GIY-YIG N-terminal endonuclease domain of UvrC from Thermotoga maritima bound to its catalytic divalent cation: manganese | | 分子名称: | GLYCEROL, MANGANESE (II) ION, UvrABC system protein C | | 著者 | Truglio, J.J, Rhau, B, Croteau, D.L, Wang, L, Skorvaga, M, Karakas, E, DellaVecchia, M.J, Wang, H, Van Houten, B, Kisker, C. | | 登録日 | 2004-12-23 | | 公開日 | 2005-03-01 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Structural insights into the first incision reaction during nucleotide excision repair
Embo J., 24, 2005
|
|
4X1T
 
 | | The crystal structure of Arabidopsis thaliana galactolipid synthase MGD1 in complex with UDP | | 分子名称: | 1,2-ETHANEDIOL, Monogalactosyldiacylglycerol synthase 1, chloroplastic, ... | | 著者 | Rocha, J, Breton, C. | | 登録日 | 2014-11-25 | | 公開日 | 2016-02-10 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Structural insights and membrane binding properties of MGD1, the major galactolipid synthase in plants.
Plant J., 85, 2016
|
|
4Q2G
 
 | | CRYSTAL STRUCTURE OF AN INTRAMEMBRANE CDP-DAG SYNTHETASE CENTRAL FOR PHOSPHOLIPID BIOSYNTHESIS (S200C/S223C, inactive mutant) | | 分子名称: | MAGNESIUM ION, MERCURY (II) ION, Phosphatidate cytidylyltransferase, ... | | 著者 | Liu, X, Yin, Y, Wu, J, Liu, Z. | | 登録日 | 2014-04-08 | | 公開日 | 2014-07-02 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (3.4 Å) | | 主引用文献 | Structure and mechanism of an intramembrane liponucleotide synthetase central for phospholipid biosynthesis
Nat Commun, 5, 2014
|
|
4JAU
 
 | | Structural basis of a rationally rewired protein-protein interface (HK853mutant A268V, A271G, T275M, V294T and D297E) | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Histidine kinase | | 著者 | Podgornaia, A.I, Casino, P, Marina, A, Laub, M.T. | | 登録日 | 2013-02-19 | | 公開日 | 2013-09-04 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Structural basis of a rationally rewired protein-protein interface critical to bacterial signaling
Structure, 21, 2013
|
|
4JXC
 
 | | X-ray snapshots of possible intermediates in the time course of synthesis and degradation of protein-bound Fe4S4 clusters | | 分子名称: | CHAPSO, CHLORIDE ION, FE2/S2 (INORGANIC) CLUSTER, ... | | 著者 | Nicolet, Y, Rohac, R, Martin, L, Fontecilla-Camps, J.C. | | 登録日 | 2013-03-28 | | 公開日 | 2013-05-01 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | X-ray snapshots of possible intermediates in the time course of synthesis and degradation of protein-bound Fe4S4 clusters.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4JAS
 
 | | Structural basis of a rationally rewired protein-protein interface (HK853mutant A268V, A271G, T275M, V294T and D297E and RR468mutant V13P, L14I, I17M and N21V) | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Histidine kinase, MAGNESIUM ION, ... | | 著者 | Podgornaia, A.I, Casino, P, Marina, A, Laub, M.T. | | 登録日 | 2013-02-19 | | 公開日 | 2013-09-04 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Structural basis of a rationally rewired protein-protein interface critical to bacterial signaling
Structure, 21, 2013
|
|
4JA2
 
 | | Structural basis of a rationally rewired protein-protein interface (RR468mutant V13P, L14I, I17M and N21V) | | 分子名称: | ACETATE ION, MAGNESIUM ION, Response regulator, ... | | 著者 | Podgornaia, A.I, Casino, P, Marina, A, Laub, M.T. | | 登録日 | 2013-02-18 | | 公開日 | 2013-09-04 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.79 Å) | | 主引用文献 | Structural basis of a rationally rewired protein-protein interface critical to bacterial signaling
Structure, 21, 2013
|
|
4JY8
 
 | | X-ray snapshots of possible intermediates in the time course of synthesis and degradation of protein-bound Fe4S4 clusters | | 分子名称: | CHLORIDE ION, FEFE-HYDROGENASE MATURASE, HYDROSULFURIC ACID, ... | | 著者 | Nicolet, Y, Rohac, R, Martin, L, Fontecilla-Camps, J.C. | | 登録日 | 2013-03-29 | | 公開日 | 2013-05-01 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.901 Å) | | 主引用文献 | X-ray snapshots of possible intermediates in the time course of synthesis and degradation of protein-bound Fe4S4 clusters.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4JAV
 
 | | Structural basis of a rationally rewired protein-protein interface (HK853wt and RR468mutant V13P, L14I, I17M and N21V) | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, Histidine kinase, ... | | 著者 | Podgornaia, A.I, Casino, P, Marina, A, Laub, M.T. | | 登録日 | 2013-02-19 | | 公開日 | 2013-09-04 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Structural basis of a rationally rewired protein-protein interface critical to bacterial signaling
Structure, 21, 2013
|
|
4JY9
 
 | | X-ray snapshots of possible intermediates in the time course of synthesis and degradation of protein-bound Fe4S4 clusters | | 分子名称: | CHAPSO, CHLORIDE ION, FE2/S2 (INORGANIC) CLUSTER, ... | | 著者 | Nicolet, Y, Rohac, R, Martin, L, Fontecilla-Camps, J.C. | | 登録日 | 2013-03-29 | | 公開日 | 2013-05-01 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | X-ray snapshots of possible intermediates in the time course of synthesis and degradation of protein-bound Fe4S4 clusters.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4JYF
 
 | | X-ray snapshots of possible intermediates in the time course of synthesis and degradation of protein-bound Fe4S4 clusters. | | 分子名称: | CARBONATE ION, CHAPSO, CHLORIDE ION, ... | | 著者 | Nicolet, Y, Rohac, R, Martin, L, Fontecilla-Camps, J.C. | | 登録日 | 2013-03-29 | | 公開日 | 2013-05-01 | | 最終更新日 | 2013-05-15 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | X-ray snapshots of possible intermediates in the time course of synthesis and degradation of protein-bound Fe4S4 clusters.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4JYE
 
 | | X-ray snapshots of possible intermediates in the time course of synthesis and degradation of protein-bound Fe4S4 clusters. | | 分子名称: | BROMIDE ION, Biotin synthetase, putative, ... | | 著者 | Nicolet, Y, Rohac, R, Martin, L, Fontecilla-Camps, J.C. | | 登録日 | 2013-03-29 | | 公開日 | 2013-05-01 | | 最終更新日 | 2013-05-15 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | X-ray snapshots of possible intermediates in the time course of synthesis and degradation of protein-bound Fe4S4 clusters.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4JYD
 
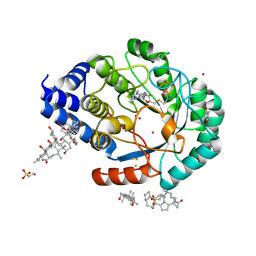 | | X-ray snapshots of possible intermediates in the time course of synthesis and degradation of protein-bound Fe4S4 clusters. | | 分子名称: | 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, BROMIDE ION, CHAPSO, ... | | 著者 | Nicolet, Y, Rohac, R, Martin, L, Fontecilla-Camps, J.C. | | 登録日 | 2013-03-29 | | 公開日 | 2013-05-01 | | 最終更新日 | 2013-05-15 | | 実験手法 | X-RAY DIFFRACTION (1.71 Å) | | 主引用文献 | X-ray snapshots of possible intermediates in the time course of synthesis and degradation of protein-bound Fe4S4 clusters.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
5H72
 
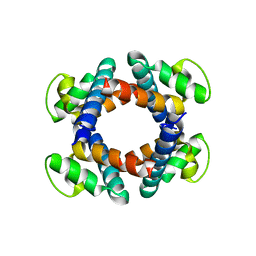 | | Structure of the periplasmic domain of FliP | | 分子名称: | Flagellar biosynthetic protein FliP | | 著者 | Fukumura, T, Kawaguchi, T, Saijo-Hamano, Y, Namba, K, Minamino, T, Imada, K. | | 登録日 | 2016-11-16 | | 公開日 | 2017-08-02 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Assembly and stoichiometry of the core structure of the bacterial flagellar type III export gate complex
PLoS Biol., 15, 2017
|
|
5HM4
 
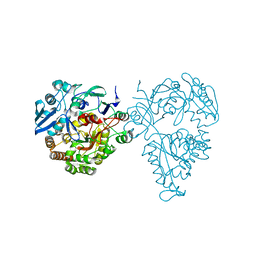 | | Crystal structure of oligopeptide ABC transporter, periplasmic oligopeptide-binding protein (TM1226) from THERMOTOGA MARITIMA at 2.0 A resolution | | 分子名称: | CALCIUM ION, Mannoside ABC transport system, sugar-binding protein | | 著者 | Lu, X, Ghimire-Rijal, S, Myles, D.A.A, Cuneo, M.J. | | 登録日 | 2016-01-15 | | 公開日 | 2016-11-23 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Periplasmic Binding Protein Dimer Has a Second Allosteric Event Tied to Ligand Binding.
Biochemistry, 56, 2017
|
|
3GTY
 
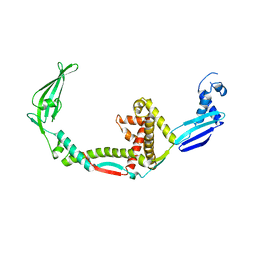 | |
3H3A
 
 | |
1Q7Q
 
 | | Cobalamin-dependent methionine synthase (1-566) from T. maritima (Oxidized, Orthorhombic) | | 分子名称: | 5-methyltetrahydrofolate S-homocysteine methyltransferase | | 著者 | Evans, J.C, Huddler, D.P, Hilgers, M.T, Romanchuk, G, Matthews, R.G, Ludwig, M.L. | | 登録日 | 2003-08-19 | | 公開日 | 2004-03-23 | | 最終更新日 | 2017-10-11 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Structures of the N-terminal modules imply large domain motions during catalysis by methionine synthase.
Proc.Natl.Acad.Sci.Usa, 101, 2004
|
|
3H38
 
 | |
3H37
 
 | |
3H39
 
 | |
3GU0
 
 | |
6DTT
 
 | | Apo T. maritima MalE2 | | 分子名称: | maltose-binding protein MalE2 | | 著者 | Cuneo, M.J, Shukla, S. | | 登録日 | 2018-06-18 | | 公開日 | 2018-09-19 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Differential Substrate Recognition by Maltose Binding Proteins Influenced by Structure and Dynamics.
Biochemistry, 57, 2018
|
|
6DTU
 
 | | Maltotetraose bound T. maritima MalE1 | | 分子名称: | alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, maltose-binding protein MalE1 | | 著者 | Cuneo, M.J, Shukla, S. | | 登録日 | 2018-06-18 | | 公開日 | 2018-09-19 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Differential Substrate Recognition by Maltose Binding Proteins Influenced by Structure and Dynamics.
Biochemistry, 57, 2018
|
|
