6T2M
 
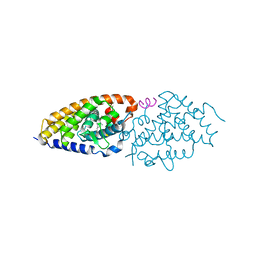 | | VDR-ZK168281 complex | | 分子名称: | Nuclear receptor coactivator 1, Vitamin D3 receptor A, ethyl (~{Z})-3-[1-[(~{E},1~{R},4~{R})-4-[(1~{R},3~{a}~{S},4~{E},7~{a}~{R})-7~{a}-methyl-4-[(2~{Z})-2-[(3~{S},5~{R})-2-methylidene-3,5-bis(oxidanyl)cyclohexylidene]ethylidene]-2,3,3~{a},5,6,7-hexahydro-1~{H}-inden-1-yl]-1-oxidanyl-pent-2-enyl]cyclopropyl]prop-2-enoate | | 著者 | Rochel, N, Belorusova, A.Y. | | 登録日 | 2019-10-09 | | 公開日 | 2020-08-26 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Structural Analysis of VDR Complex with ZK168281 Antagonist.
J.Med.Chem., 63, 2020
|
|
2DA6
 
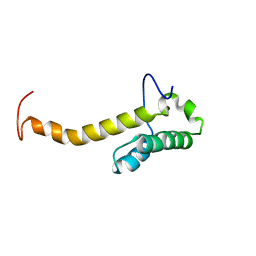 | | Solution structure of the homeobox domain of Hepatocyte nuclear factor 1-beta (HNF-1beta) | | 分子名称: | Hepatocyte nuclear factor 1-beta | | 著者 | Ohnishi, S, Kigawa, T, Sato, M, Koshiba, S, Inoue, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2005-12-13 | | 公開日 | 2006-12-26 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of the homeobox domain of Hepatocyte nuclear factor 1-beta (HNF-1beta)
To be Published
|
|
1V4F
 
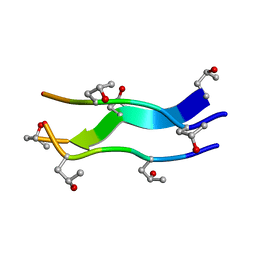 | | Crystal structures of collagen model peptides with pro-hyp-gly sequence at 1.3A | | 分子名称: | collagen like peptide | | 著者 | Okuyama, K, Hongo, C, Fukushima, R, Wu, G, Noguchi, K, Tanaka, Y, Nishino, N. | | 登録日 | 2003-11-13 | | 公開日 | 2004-08-03 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.26 Å) | | 主引用文献 | Crystal structures of collagen model peptides with Pro-Hyp-Gly repeating sequence at 1.26 A resolution: implications for proline ring puckering
Biopolymers, 76, 2004
|
|
1V5K
 
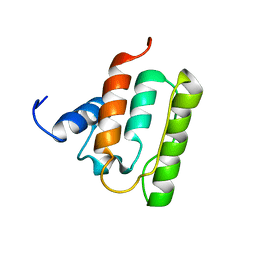 | | Solution structure of the CH domain from mouse EB-1 | | 分子名称: | microtubule-associated protein, RP/EB family, member 1 | | 著者 | Tomizawa, T, Kigawa, T, Koshiba, S, Inoue, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2003-11-25 | | 公開日 | 2004-05-25 | | 最終更新日 | 2023-12-27 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of the CH domain from mouse EB-1
To be Published
|
|
1V7H
 
 | | Crystal Structures of Collagen Model Peptides with Pro-Hyp-Gly Sequence at 1.26 A | | 分子名称: | Collagen like peptide | | 著者 | Okuyama, K, Hongo, C, Fukushima, R, Wu, G, Narita, H, Noguchi, K, Tanaka, Y, Nishino, N. | | 登録日 | 2003-12-17 | | 公開日 | 2004-08-03 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.25 Å) | | 主引用文献 | Crystal structures of collagen model peptides with Pro-Hyp-Gly repeating sequence at 1.26 A resolution: implications for proline ring puckering
Biopolymers, 76, 2004
|
|
2DAD
 
 | | Solution structure of the fifth crystall domain of the non-lens protein, Absent in melanoma 1 | | 分子名称: | Absent in melanoma 1 protein | | 著者 | Ohnishi, S, Kigawa, T, Saito, K, Koshiba, S, Inoue, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2005-12-13 | | 公開日 | 2006-06-13 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of the fifth crystall domain of the non-lens protein, Absent in melanoma 1
To be Published
|
|
1VA0
 
 | | Crystal Structure of the Native Form of Uroporphyrin III C-methyl transferase from Thermus thermophilus | | 分子名称: | 1,2-ETHANEDIOL, CHLORIDE ION, Uroporphyrin-III C-methyltransferase | | 著者 | Rehse, P.H, Kitao, T, Tahirov, T.H, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2004-02-05 | | 公開日 | 2005-02-15 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.97 Å) | | 主引用文献 | Structure of a closed-form uroporphyrinogen-III C-methyltransferase from Thermus thermophilus.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
7G0B
 
 | | Crystal Structure of human FABP5 in complex with 7-bromo-1-methyl-5-phenyl-2,3,4,5-tetrahydro-1-benzazepine-4-carboxylic acid, i.e. SMILES [C@@H]1([C@@H](CCN(c2c1cc(cc2)Br)C)C(=O)O)c1ccccc1 with IC50=2.3 microM | | 分子名称: | (4S,5S)-7-bromo-1-methyl-5-phenyl-2,3,4,5-tetrahydro-1H-1-benzazepine-4-carboxylic acid, DIMETHYL SULFOXIDE, Fatty acid-binding protein 5, ... | | 著者 | Ehler, A, Benz, J, Obst, U, Ceccarelli-Simona, M, Rudolph, M.G. | | 登録日 | 2023-04-27 | | 公開日 | 2023-06-14 | | 最終更新日 | 2025-08-13 | | 実験手法 | X-RAY DIFFRACTION (1.47 Å) | | 主引用文献 | A high-resolution data set of fatty acid-binding protein structures. III. Unexpectedly high occurrence of wrong ligands.
Acta Crystallogr D Struct Biol, 81, 2025
|
|
7FWI
 
 | | Crystal Structure of human FABP5 in complex with 2-(indole-1-carbonylamino)benzoic acid, i.e. SMILES c12N(C(=O)Nc3c(cccc3)C(=O)O)C=Cc1cccc2 with IC50=18.1696 microM | | 分子名称: | 2-[(2,3-dihydro-1H-indole-1-carbonyl)amino]benzoic acid, CHLORIDE ION, Fatty acid-binding protein 5 | | 著者 | Ehler, A, Benz, J, Obst, U, Ning, R, Rudolph, M.G. | | 登録日 | 2023-04-27 | | 公開日 | 2023-06-14 | | 最終更新日 | 2025-08-13 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | A high-resolution data set of fatty acid-binding protein structures. III. Unexpectedly high occurrence of wrong ligands.
Acta Crystallogr D Struct Biol, 81, 2025
|
|
7FX5
 
 | | Crystal Structure of human FABP4 in complex with 5-[[3-(3-cyclopropyl-1,2,4-oxadiazol-5-yl)-6,6-difluoro-5,7-dihydro-4H-1-benzothiophen-2-yl]carbamoyl]-3,6-dihydro-2H-pyran-4-carboxylic acid, i.e. SMILES C1C(CC2=C(C1)C(=C(S2)NC(=O)C1=C(CCOC1)C(=O)O)C1=NC(=NO1)C1CC1)(F)F with IC50=0.182863 microM | | 分子名称: | 5-{[(3M)-3-(3-cyclopropyl-1,2,4-oxadiazol-5-yl)-6,6-difluoro-4,5,6,7-tetrahydro-1-benzothiophen-2-yl]carbamoyl}-3,6-dihydro-2H-pyran-4-carboxylic acid, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | 著者 | Ehler, A, Benz, J, Obst, U, Richter, H, Rudolph, M.G. | | 登録日 | 2023-04-27 | | 公開日 | 2023-06-14 | | 最終更新日 | 2025-08-13 | | 実験手法 | X-RAY DIFFRACTION (1.12 Å) | | 主引用文献 | A high-resolution data set of fatty acid-binding protein structures. III. Unexpectedly high occurrence of wrong ligands.
Acta Crystallogr D Struct Biol, 81, 2025
|
|
3ZD2
 
 | | THE STRUCTURE OF THE TWO N-TERMINAL DOMAINS OF COMPLEMENT FACTOR H RELATED PROTEIN 1 SHOWS FORMATION OF A NOVEL DIMERISATION INTERFACE | | 分子名称: | 1,2-ETHANEDIOL, COMPLEMENT FACTOR H-RELATED PROTEIN 1 | | 著者 | Caesar, J.J.E, Goicoechea de Jorge, E, Pickering, M.C, Lea, S.M. | | 登録日 | 2012-11-23 | | 公開日 | 2013-03-13 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (1.99 Å) | | 主引用文献 | Dimerization of Complement Factor H-Related Proteins Modulates Complement Activation in Vivo.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4QYF
 
 | |
6JUJ
 
 | | Crystal structure of Formate dehydrogenase mutant V198I/C256I/P260S/E261P/S381N/S383F from Pseudomonas sp. 101in complex with non-natural cofactor Nicotinamide Cytosine Dinucleotide | | 分子名称: | Formate dehydrogenase, GLYCEROL, [[(2S,3S,4R,5S)-5-(3-aminocarbonylpyridin-1-ium-1-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2S,3S,4R,5S)-5-(4-azanyl-2-oxidanylidene-pyrimidin-1-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl hydrogen phosphate | | 著者 | Feng, Y, Guo, X, Xue, S, Zhao, Z. | | 登録日 | 2019-04-14 | | 公開日 | 2020-05-13 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.183 Å) | | 主引用文献 | Structure-Guided Design of Formate Dehydrogenase for Regeneration of a Non-Natural Redox Cofactor.
Chemistry, 26, 2020
|
|
5L61
 
 | |
1PPK
 
 | |
7FZI
 
 | | Crystal Structure of human FABP4 in complex with 2-[[2-(3-cyclopropyl-1,2,4-oxadiazol-5-yl)phenyl]carbamoyl]cyclopentene-1-carboxylic acid, i.e. SMILES N(c1ccccc1C1=NC(=NO1)C1CC1)C(=O)C1=C(CCC1)C(=O)O with IC50=0.0623077 microM | | 分子名称: | 2-{[(2M)-2-(3-cyclopropyl-1,2,4-oxadiazol-5-yl)phenyl]carbamoyl}cyclopent-1-ene-1-carboxylic acid, DIMETHYL SULFOXIDE, Fatty acid-binding protein, ... | | 著者 | Ehler, A, Benz, J, Obst, U, Richter, H, Rudolph, M.G. | | 登録日 | 2023-04-27 | | 公開日 | 2023-06-14 | | 最終更新日 | 2025-08-13 | | 実験手法 | X-RAY DIFFRACTION (1.07 Å) | | 主引用文献 | A high-resolution data set of fatty acid-binding protein structures. III. Unexpectedly high occurrence of wrong ligands.
Acta Crystallogr D Struct Biol, 81, 2025
|
|
7FZW
 
 | | Crystal Structure of human FABP4 in complex with 2-[[5-bromo-2-(3-cyclopropyl-1,2,4-oxadiazol-5-yl)phenyl]carbamoyl]cyclopentene-1-carboxylic acid, i.e. SMILES C1(=C(CCC1)C(=O)O)C(=O)Nc1cc(ccc1C1=NC(=NO1)C1CC1)Br with IC50=0.333165 microM | | 分子名称: | 2-{[(2M)-5-bromo-2-(3-cyclopropyl-1,2,4-oxadiazol-5-yl)phenyl]carbamoyl}cyclopent-1-ene-1-carboxylic acid, BROMIDE ION, DIMETHYL SULFOXIDE, ... | | 著者 | Ehler, A, Benz, J, Obst, U, Richter, H, Rudolph, M.G. | | 登録日 | 2023-04-27 | | 公開日 | 2023-06-14 | | 最終更新日 | 2025-08-13 | | 実験手法 | X-RAY DIFFRACTION (1.24 Å) | | 主引用文献 | A high-resolution data set of fatty acid-binding protein structures. III. Unexpectedly high occurrence of wrong ligands.
Acta Crystallogr D Struct Biol, 81, 2025
|
|
4R93
 
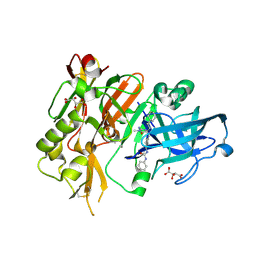 | | BACE-1 in complex with (R)-4-(2-cyclohexylethyl)-1-methyl-5-oxo-4-(((1S,3R)-3-(3-phenylureido)cyclohexyl)methyl)imidazolidin-2-iminium | | 分子名称: | 1-[(1R,3S)-3-{[(2E,4R)-4-(2-cyclohexylethyl)-2-imino-1-methyl-5-oxoimidazolidin-4-yl]methyl}cyclohexyl]-3-phenylurea, Beta-secretase 1, L(+)-TARTARIC ACID | | 著者 | Orth, P, Strickland, C, Caldwell, J.P. | | 登録日 | 2014-09-03 | | 公開日 | 2014-11-05 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (1.71 Å) | | 主引用文献 | Discovery of potent iminoheterocycle BACE1 inhibitors.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
1LPO
 
 | |
5RE8
 
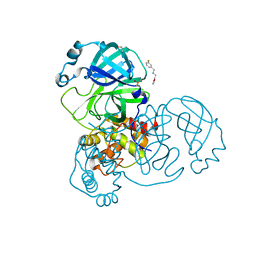 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 main protease in complex with Z2737076969 | | 分子名称: | 1-(3-fluorophenyl)-N-[(furan-2-yl)methyl]methanamine, 3C-like proteinase, DIMETHYL SULFOXIDE | | 著者 | Fearon, D, Owen, C.D, Douangamath, A, Lukacik, P, Powell, A.J, Strain-Damerell, C.M, Resnick, E, Krojer, T, Gehrtz, P, Wild, C, Aimon, A, Brandao-Neto, J, Carbery, A, Dunnett, L, Skyner, R, Snee, M, London, N, Walsh, M.A, von Delft, F. | | 登録日 | 2020-03-15 | | 公開日 | 2020-03-25 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.81 Å) | | 主引用文献 | Crystallographic and electrophilic fragment screening of the SARS-CoV-2 main protease.
Nat Commun, 11, 2020
|
|
4BV3
 
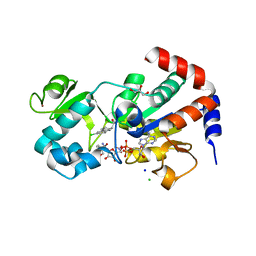 | | CRYSTAL STRUCTURE OF SIRT3 IN COMPLEX WITH THE INHIBITOR EX-527 AND NAD | | 分子名称: | (1S)-6-chloro-2,3,4,9-tetrahydro-1H-carbazole-1- carboxamide, ADENOSINE-5-DIPHOSPHORIBOSE, CHLORIDE ION, ... | | 著者 | Gertz, M, Nguyen, N.T.T, Weyand, M, Steegborn, C. | | 登録日 | 2013-06-24 | | 公開日 | 2013-07-17 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Ex-527 Inhibits Sirtuins by Exploiting Their Unique Nad+-Dependent Deacetylation Mechanism
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4BW5
 
 | | Crystal structure of human two pore domain potassium ion channel TREK2 (K2P10.1) | | 分子名称: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CADMIUM ION, POTASSIUM CHANNEL SUBFAMILY K MEMBER 10, ... | | 著者 | Pike, A.C.W, Dong, Y.Y, Dong, L, Quigley, A, Shrestha, L, Mukhopadhyay, S, Strain-Damerell, C, Goubin, S, Grieben, M, Shintre, C.A, Mackenzie, A, Vollmar, M, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Burgess-Brown, N, Carpenter, E.P. | | 登録日 | 2013-06-30 | | 公開日 | 2013-07-31 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | K2P Channel Gating Mechanisms Revealed by Structures of Trek-2 and a Complex with Prozac
Science, 347, 2015
|
|
7FZL
 
 | | Crystal Structure of human FABP4 in complex with 3-(3-chlorophenyl)-4-prop-2-enyl-1H-1,2,4-triazole-5-thione, i.e. SMILES N1(C(=NNC1=S)c1cc(Cl)ccc1)CC=C with IC50=1.4 microM | | 分子名称: | (5P)-5-(3-chlorophenyl)-4-(prop-2-en-1-yl)-2,4-dihydro-3H-1,2,4-triazole-3-thione, FORMIC ACID, Fatty acid-binding protein, ... | | 著者 | Ehler, A, Benz, J, Obst, U, Brunner, M, Rudolph, M.G. | | 登録日 | 2023-04-27 | | 公開日 | 2023-06-14 | | 最終更新日 | 2025-08-13 | | 実験手法 | X-RAY DIFFRACTION (1.12 Å) | | 主引用文献 | A high-resolution data set of fatty acid-binding protein structures. III. Unexpectedly high occurrence of wrong ligands.
Acta Crystallogr D Struct Biol, 81, 2025
|
|
2W0V
 
 | | Crystal structure of Glmu from Haemophilus influenzae in complex with quinazoline inhibitor 1 | | 分子名称: | 6-(CYCLOPROP-2-EN-1-YLMETHOXY)-2-[6-(CYCLOPROPYLMETHYL)-5-OXO-3,4,5,6-TETRAHYDRO-2,6-NAPHTHYRIDIN-2(1H)-YL]-7-METHOXYQUINAZOLIN-4(3H)-ONE, GLUCOSAMINE-1-PHOSPHATE N-ACETYLTRANSFERASE, SULFATE ION, ... | | 著者 | Mochalkin, I, Melnick, M. | | 登録日 | 2008-10-10 | | 公開日 | 2009-11-17 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.99 Å) | | 主引用文献 | Discovery and Initial Sar of Quinazoline Inhibitors of Glmu from Haemophilus Influenzae
To be Published
|
|
1DGM
 
 | | CRYSTAL STRUCTURE OF ADENOSINE KINASE FROM TOXOPLASMA GONDII | | 分子名称: | ACETIC ACID, ADENOSINE, ADENOSINE KINASE, ... | | 著者 | Cook, W.J, DeLucas, L.J, Chattopadhyay, D. | | 登録日 | 1999-11-24 | | 公開日 | 2000-11-29 | | 最終更新日 | 2024-11-13 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal structure of adenosine kinase from Toxoplasma gondii at 1.8 A resolution.
Protein Sci., 9, 2000
|
|
