3HG9
 
 | | CRYSTAL STRUCTURE OF putative pilM protein from Pseudomonas aeruginosa 2192 | | 分子名称: | NICKEL (II) ION, PilM | | 著者 | Malashkevich, V.N, Toro, R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | 登録日 | 2009-05-13 | | 公開日 | 2009-05-26 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | CRYSTAL STRUCTURE OF putative pilM protein from Pseudomonas aeruginosa 2192
To be Published
|
|
4O8H
 
 | | 0.85A resolution structure of PEG 400 Bound Cyclophilin D | | 分子名称: | DI(HYDROXYETHYL)ETHER, PENTAETHYLENE GLYCOL, Peptidyl-prolyl cis-trans isomerase F, ... | | 著者 | Lovell, S, Valasani, K.R, Battaile, K.P, Wang, C, Yan, S.S. | | 登録日 | 2013-12-27 | | 公開日 | 2014-06-11 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (0.85 Å) | | 主引用文献 | High-resolution crystal structures of two crystal forms of human cyclophilin D in complex with PEG 400 molecules.
Acta Crystallogr F Struct Biol Commun, 70, 2014
|
|
3HJ4
 
 | | Minor Editosome-Associated TUTase 1 | | 分子名称: | Minor Editosome-Associated TUTase | | 著者 | Stagno, J, Luecke, H. | | 登録日 | 2009-05-20 | | 公開日 | 2010-05-12 | | 最終更新日 | 2017-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.56 Å) | | 主引用文献 | Structure of the Mitochondrial Editosome-Like Complex Associated TUTase 1 Reveals Divergent Mechanisms of UTP Selection and Domain Organization.
J.Mol.Biol., 399, 2010
|
|
5DI9
 
 | |
4OB8
 
 | | Crystal structure of a novel thermostable esterase from Pseudomonas putida ECU1011 | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, Alpha/beta hydrolase fold-3 domain protein, DI(HYDROXYETHYL)ETHER | | 著者 | Dou, S, Kong, X.D, Ma, B.D, Xu, J.H, Zhou, J.H. | | 登録日 | 2014-01-07 | | 公開日 | 2014-07-23 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.801 Å) | | 主引用文献 | Crystal structures of Pseudomonas putida esterase reveal the functional role of residues 187 and 287 in substrate binding and chiral recognition
Biochem.Biophys.Res.Commun., 446, 2014
|
|
3HDU
 
 | |
3HER
 
 | | Human prion protein variant F198S with V129 | | 分子名称: | CADMIUM ION, Major prion protein | | 著者 | Lee, S, Antony, L, Hartmann, R, Knaus, K.J, Surewicz, K, Surewicz, W.K, Yee, V.C. | | 登録日 | 2009-05-10 | | 公開日 | 2010-01-12 | | 最終更新日 | 2021-10-13 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Conformational diversity in prion protein variants influences intermolecular beta-sheet formation.
Embo J., 29, 2010
|
|
3HJX
 
 | | Human prion protein variant D178N with V129 | | 分子名称: | CADMIUM ION, CHLORIDE ION, Major prion protein | | 著者 | Lee, S, Antony, L, Hartmann, R, Knaus, K.J, Surewicz, K, Surewicz, W.K, Yee, V.C. | | 登録日 | 2009-05-22 | | 公開日 | 2010-01-12 | | 最終更新日 | 2021-10-13 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Conformational diversity in prion protein variants influences intermolecular beta-sheet formation.
Embo J., 29, 2010
|
|
3HGY
 
 | |
4OEM
 
 | | Crystal structure of Cathepsin C in complex with dipeptide substrates | | 分子名称: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | 著者 | Zhao, B, Smallwood, A, Concha, N. | | 登録日 | 2014-01-13 | | 公開日 | 2015-03-25 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (1.52 Å) | | 主引用文献 | The amino-acid substituents of dipeptide substrates of cathepsin C can determine the rate-limiting steps of catalysis.
Biochemistry, 51, 2012
|
|
3HHP
 
 | |
3HK1
 
 | | Identification and Characterization of a Small Molecule Inhibitor of Fatty Acid Binding Proteins | | 分子名称: | 4-{[2-(methoxycarbonyl)-5-(2-thienyl)-3-thienyl]amino}-4-oxo-2-butenoic acid, Fatty acid-binding protein, adipocyte | | 著者 | Hertzel, A.V, Hellberg, K, Reynolds, J.M, Kruse, A.C, Juhlmann, B.E, Smith, A.J, Sanders, M.A, Ohlendorf, D.H, Suttles, J, Bernlohr, D.A. | | 登録日 | 2009-05-22 | | 公開日 | 2009-09-22 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Identification and characterization of a small molecule inhibitor of Fatty Acid binding proteins.
J.Med.Chem., 52, 2009
|
|
4OB7
 
 | | Crystal structure of esterase rPPE mutant W187H | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, Alpha/beta hydrolase fold-3 domain protein, DI(HYDROXYETHYL)ETHER | | 著者 | Dou, S, Kong, X.D, Ma, B.D, Xu, J.H, Zhou, J.H. | | 登録日 | 2014-01-07 | | 公開日 | 2014-07-23 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Crystal structures of Pseudomonas putida esterase reveal the functional role of residues 187 and 287 in substrate binding and chiral recognition
Biochem.Biophys.Res.Commun., 446, 2014
|
|
3HEF
 
 | |
3HFC
 
 | | A trimeric form of the Kv7.1 A domain Tail, L602M/L606M mutant Semet | | 分子名称: | Potassium voltage-gated channel subfamily KQT member 1 | | 著者 | Xu, Q, Minor, D.L. | | 登録日 | 2009-05-11 | | 公開日 | 2009-09-01 | | 最終更新日 | 2021-10-13 | | 実験手法 | X-RAY DIFFRACTION (2.45 Å) | | 主引用文献 | Crystal structure of a trimeric form of the K(V)7.1 (KCNQ1) A-domain tail coiled-coil reveals structural plasticity and context dependent changes in a putative coiled-coil trimerization motif.
Protein Sci., 18, 2009
|
|
3HK4
 
 | |
3HG6
 
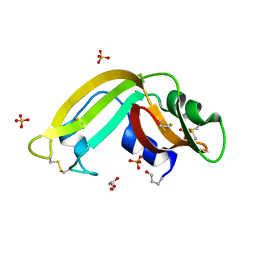 | | Crystal Structure of the Recombinant Onconase from Rana pipiens | | 分子名称: | GLYCEROL, Onconase, SULFATE ION | | 著者 | Camara-Artigas, A, Gavira, J.A, Casares-Atienza, S, Weininger, U, Balbach, J, Garcia-Mira, M.M. | | 登録日 | 2009-05-13 | | 公開日 | 2010-05-19 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Three-state thermal unfolding of onconase.
Biophys.Chem., 159, 2011
|
|
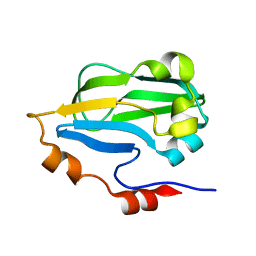
3HGB
 
 | |
4OM8
 
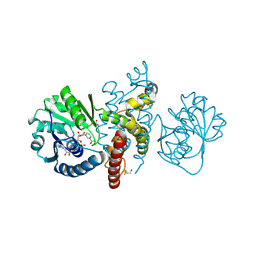 | | Crystal structure of 5-formly-3-hydroxy-2-methylpyridine 4-carboxylic acid (FHMPC) 5-dehydrogenase, an NAD+ dependent dismutase. | | 分子名称: | 3-hydroxybutyryl-coA dehydrogenase, ACETATE ION, BETA-MERCAPTOETHANOL, ... | | 著者 | Mugo, A.N, Kobayashi, J, Mikami, B, Yagi, T, Ohnishi, K. | | 登録日 | 2014-01-27 | | 公開日 | 2015-01-28 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Crystal structure of 5-formyl-3-hydroxy-2-methylpyridine 4-carboxylic acid 5-dehydrogenase, an NAD(+)-dependent dismutase from Mesorhizobium loti
Biochem.Biophys.Res.Commun., 456, 2015
|
|
6QMU
 
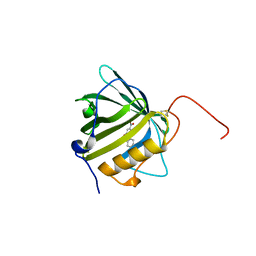 | |
3HIS
 
 | |
7M7W
 
 | | Antibodies to the SARS-CoV-2 receptor-binding domain that maximize breadth and resistance to viral escape | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Monoclonal antibody S2H97 Fab heavy chain, Monoclonal antibody S2H97 Fab light chain, ... | | 著者 | Snell, G, Czudnochowski, N, Croll, T.I, Nix, J.C, Corti, D, Cameroni, E, Pinto, D, Beltramello, M. | | 登録日 | 2021-03-29 | | 公開日 | 2021-05-05 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.65 Å) | | 主引用文献 | SARS-CoV-2 RBD antibodies that maximize breadth and resistance to escape.
Nature, 597, 2021
|
|
4OH7
 
 | |
6QNR
 
 | | 70S ribosome elongation complex (EC) with experimentally assigned potassium ions | | 分子名称: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | 著者 | Rozov, A, Khusainov, I, Yusupov, M, Yusupova, G. | | 登録日 | 2019-02-11 | | 公開日 | 2019-06-19 | | 最終更新日 | 2024-04-24 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Importance of potassium ions for ribosome structure and function revealed by long-wavelength X-ray diffraction.
Nat Commun, 10, 2019
|
|
4OOV
 
 | |
