7SDH
 
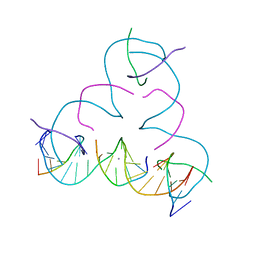 | | [S:Ag+:S] Metal-mediated DNA base pair in a self-assembling rhombohedral lattice | | 分子名称: | DNA (5'-D(*GP*AP*GP*CP*AP*GP*CP*CP*TP*GP*TP*(IMC)P*TP*GP*GP*AP*CP*AP*TP*CP*A)-3'), DNA (5'-D(P*CP*CP*AP*(IMC)P*AP*CP*A)-3'), DNA (5'-D(P*CP*TP*GP*AP*TP*GP*T)-3'), ... | | 著者 | Lu, B, Vecchioni, S, Seeman, N.C, Sha, R, Ohayon, Y.P. | | 登録日 | 2021-09-29 | | 公開日 | 2022-10-05 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (4.05 Å) | | 主引用文献 | Metal-Mediated DNA Nanotechnology in 3D: Structural Library by Templated Diffraction.
Adv Mater, 2023
|
|
6ZUQ
 
 | | Crystal structure of the effector Ecp11-1 from Fulvia fulva | | 分子名称: | Extracellular protein 11-1, GLYCEROL, ZINC ION | | 著者 | Lazar, N, Mesarich, C, Petit-Houdenot, Y, Talbi, N, Li de la Sierra-Gallay, I, Zelie, E, Blondeau, K, Gracy, J, Ollivier, B, van de Wouw, A, Balesdent, M.H, Idnurm, A, van Tilbeurgh, H, Fudal, I. | | 登録日 | 2020-07-23 | | 公開日 | 2021-08-04 | | 最終更新日 | 2022-07-27 | | 実験手法 | X-RAY DIFFRACTION (1.94 Å) | | 主引用文献 | A new family of structurally conserved fungal effectors displays epistatic interactions with plant resistance proteins.
Plos Pathog., 18, 2022
|
|
7SDW
 
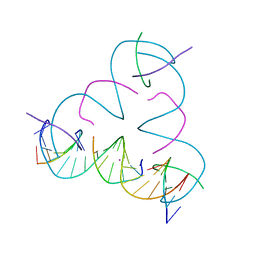 | | [T:Hg2+:mC] Metal-mediated DNA base pair in a self-assembling rhombohedral lattice | | 分子名称: | DNA (5'-D(*GP*AP*GP*CP*AP*GP*CP*CP*TP*GP*TP*TP*TP*GP*GP*AP*CP*AP*TP*CP*A)-3'), DNA (5'-D(P*CP*CP*AP*(5CM)P*AP*CP*A)-3'), DNA (5'-D(P*CP*TP*GP*AP*TP*GP*T)-3'), ... | | 著者 | Lu, B, Vecchioni, S, Seeman, N.C, Sha, R, Ohayon, Y.P. | | 登録日 | 2021-09-29 | | 公開日 | 2022-10-05 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (4.2 Å) | | 主引用文献 | Metal-Mediated DNA Nanotechnology in 3D: Structural Library by Templated Diffraction.
Adv Mater, 2023
|
|
7PNG
 
 | |
8OY3
 
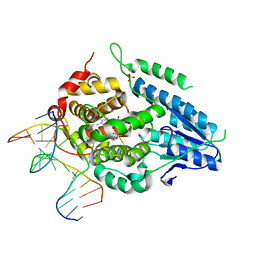 | | Time-resolved SFX structure of the class II photolyase complexed with a thymine dimer (3 picosecond pump-probe delay) | | 分子名称: | COUNTERSTRAND-OLIGONUCLEOTIDE, CPD-COMPRISING OLIGONUCLEOTIDE, DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, ... | | 著者 | Lane, T.J, Christou, N.-E, Melo, D.V.M, Apostolopoulou, V, Pateras, A, Mashhour, A.R, Galchenkova, M, Gunther, S, Reinke, P, Kremling, V, Oberthuer, D, Henkel, A, Sprenger, J, Scheer, T.E.S, Lange, E, Yefanov, O.N, Middendorf, P, Sellberg, J.A, Schubert, R, Fadini, A, Cirelli, C, Beale, E.V, Johnson, P, Dworkowski, F, Ozerov, D, Bertrand, Q, Wranik, M, Zitter, E.D, Turk, D, Bajt, S, Chapman, H, Bacellar, C. | | 登録日 | 2023-05-03 | | 公開日 | 2023-11-22 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.16 Å) | | 主引用文献 | Time-resolved crystallography captures light-driven DNA repair.
Science, 382, 2023
|
|
2YZG
 
 | |
7SG8
 
 | | [T-T] DNA mismatch in a self-assembling rhombohedral lattice | | 分子名称: | DNA (5'-D(*GP*AP*GP*CP*AP*GP*CP*CP*TP*GP*TP*TP*TP*GP*GP*AP*CP*AP*TP*CP*A)-3'), DNA (5'-D(P*CP*CP*AP*TP*AP*CP*A)-3'), DNA (5'-D(P*CP*TP*GP*AP*TP*GP*T)-3'), ... | | 著者 | Vecchioni, S, Lu, B, Bernfeld, W, Seeman, N.C, Sha, R, Ohayon, Y.P. | | 登録日 | 2021-10-05 | | 公開日 | 2022-10-19 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (3.71 Å) | | 主引用文献 | Metal-Mediated DNA Nanotechnology in 3D: Structural Library by Templated Diffraction.
Adv Mater, 2023
|
|
3GXV
 
 | | Three-dimensional structure of N-terminal domain of DnaB Helicase from Helicobacter pylori and its interactions with primase | | 分子名称: | Replicative DNA helicase | | 著者 | Kashav, T, Nitharwal, R, Syed, A.A, Gabdoulkhakov, A, Saenger, W, Dhar, K.S, Gourinath, S. | | 登録日 | 2009-04-03 | | 公開日 | 2010-01-26 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Three-dimensional structure of N-terminal domain of DnaB helicase and helicase-primase interactions in Helicobacter pylori
Plos One, 4, 2009
|
|
8P0L
 
 | | Crystal structure of human O-GlcNAcase in complex with an S-linked CKII peptide | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CYSTEINE, Protein O-GlcNAcase, ... | | 著者 | Males, A, Davies, G.J, Calvelo, M, Alteen, M.G, Vocadlo, D.J, Rovira, C. | | 登録日 | 2023-05-10 | | 公開日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Human O -GlcNAcase Uses a Preactivated Boat-skew Substrate Conformation for Catalysis. Evidence from X-ray Crystallography and QM/MM Metadynamics.
Acs Catalysis, 13, 2023
|
|
5KCV
 
 | | Crystal structure of allosteric inhibitor, ARQ 092, in complex with autoinhibited form of AKT1 | | 分子名称: | 3-[3-[4-(1-azanylcyclobutyl)phenyl]-5-phenyl-imidazo[4,5-b]pyridin-2-yl]pyridin-2-amine, RAC-alpha serine/threonine-protein kinase | | 著者 | Eathiraj, S. | | 登録日 | 2016-06-07 | | 公開日 | 2016-06-29 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Discovery of 3-(3-(4-(1-Aminocyclobutyl)phenyl)-5-phenyl-3H-imidazo[4,5-b]pyridin-2-yl)pyridin-2-amine (ARQ 092): An Orally Bioavailable, Selective, and Potent Allosteric AKT Inhibitor.
J.Med.Chem., 59, 2016
|
|
7PQ8
 
 | | Crystal structure of Campylobacter jejuni DsbA1 | | 分子名称: | TETRAETHYLENE GLYCOL, Thiol:disulfide interchange protein DsbA | | 著者 | Orlikowska, M, Bocian-Ostrzycka, K.M, Banas, A.M, Jagusztyn-Krynicka, E.K. | | 登録日 | 2021-09-16 | | 公開日 | 2021-12-29 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.329 Å) | | 主引用文献 | Interplay between DsbA1, DsbA2 and C8J_1298 Periplasmic Oxidoreductases of Campylobacter jejuni and Their Impact on Bacterial Physiology and Pathogenesis.
Int J Mol Sci, 22, 2021
|
|
7DGW
 
 | | De novo designed protein H4A2S | | 分子名称: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, de novo designed protein H4A2S | | 著者 | Xu, Y, Liao, S, Chen, Q, Liu, H. | | 登録日 | 2020-11-12 | | 公開日 | 2021-11-24 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (1.35 Å) | | 主引用文献 | A backbone-centred energy function of neural networks for protein design.
Nature, 602, 2022
|
|
6ZW1
 
 | | X-ray structure of Danio rerio histone deacetylase 6 (HDAC6) CD2 in complex with an inhibitor SW101 | | 分子名称: | 1-[[4-(oxidanylcarbamoyl)phenyl]methyl]-3,4-dihydro-2~{H}-quinoline-6-carboxamide, GLYCEROL, Histone deacetylase 6, ... | | 著者 | Barinka, C, Motlova, L, Ustinova, K. | | 登録日 | 2020-07-27 | | 公開日 | 2021-08-04 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.13 Å) | | 主引用文献 | Tetrahydroquinoline-Capped Histone Deacetylase 6 Inhibitor SW-101 Ameliorates Pathological Phenotypes in a Charcot-Marie-Tooth Type 2A Mouse Model.
J.Med.Chem., 64, 2021
|
|
5KDI
 
 | | How FAPP2 Selects Simple Glycosphingolipids Using the GLTP-fold | | 分子名称: | (~{Z})-~{N}-[(~{E},2~{S},3~{R})-1-[(2~{R},3~{R},4~{S},5~{R},6~{R})-6-(hydroxymethyl)-3,4,5-tris(oxidanyl)oxan-2-yl]oxy-3-oxidanyl-octadec-4-en-2-yl]octadec-9-enamide, Pleckstrin homology domain-containing family A member 8 | | 著者 | Ochoa-Lizarralde, B, Popov, A.N, Samygina, V.R, Patel, D.J, Brown, R.E, Malinina, L. | | 登録日 | 2016-06-08 | | 公開日 | 2017-12-13 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Structural analyses of 4-phosphate adaptor protein 2 yield mechanistic insights into sphingolipid recognition by the glycolipid transfer protein family.
J.Biol.Chem., 293, 2018
|
|
7DJI
 
 | |
2FRH
 
 | | Crystal Structure of Sara, A Transcription Regulator From Staphylococcus Aureus | | 分子名称: | CALCIUM ION, Staphylococcal accessory regulator A | | 著者 | Liu, Y, Manna, A.C, Ingavale, S, Cheung, A.L, Zhang, G. | | 登録日 | 2006-01-19 | | 公開日 | 2006-01-31 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structural and function analyses of the global regulatory protein SarA from Staphylococcus aureus.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
8P35
 
 | | Mutant human titin immunoglobulin-like 21 domain - C3575S | | 分子名称: | Titin | | 著者 | Martinez-Martin, I, Crousilles, A, Mortensen, S.A, Alegre-Cebollada, J, Wilmanns, M. | | 登録日 | 2023-05-17 | | 公開日 | 2023-11-29 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Titin domains with reduced core hydrophobicity cause dilated cardiomyopathy.
Cell Rep, 42, 2023
|
|
7DGG
 
 | |
6ZZ2
 
 | | Cocktail experiment E: fragments 52, 58, and 63 at 90 mM concentration in complex with Endothiapepsin | | 分子名称: | DIMETHYL SULFOXIDE, Endothiapepsin, GLYCEROL, ... | | 著者 | Hassaan, E, Klebe, G, Heine, A, Schiebel, J, Koester, H. | | 登録日 | 2020-08-03 | | 公開日 | 2021-08-11 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.14885437 Å) | | 主引用文献 | Cocktail experiment E: fragments 52, 58, and 63 at 90 mM concentration in complex with Endothiapepsin
To Be Published
|
|
7PRG
 
 | | Joint X-ray/neutron room temperature structure of perdeuterated LecB lectin in complex with perdeuterated fucose | | 分子名称: | CALCIUM ION, Fucose-binding lectin, SULFATE ION, ... | | 著者 | Gajdos, L, Blakeley, M.P, Haertlein, M, Forsyth, T.V, Devos, J.M, Imberty, A. | | 登録日 | 2021-09-21 | | 公開日 | 2022-01-12 | | 最終更新日 | 2024-05-01 | | 実験手法 | NEUTRON DIFFRACTION (1.85 Å), X-RAY DIFFRACTION | | 主引用文献 | Neutron crystallography reveals mechanisms used by Pseudomonas aeruginosa for host-cell binding.
Nat Commun, 13, 2022
|
|
7SGK
 
 | |
2LU4
 
 | |
2FSP
 
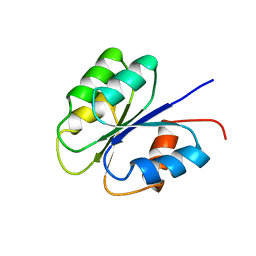 | | NMR SOLUTION STRUCTURE OF BACILLUS SUBTILIS SPO0F PROTEIN, MINIMIZED AVERAGE STRUCTURE | | 分子名称: | STAGE 0 SPORULATION PROTEIN F | | 著者 | Feher, V.A, Skelton, N.J, Dahlquist, F.W, Cavanagh, J. | | 登録日 | 1997-06-06 | | 公開日 | 1997-12-10 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | High-resolution NMR structure and backbone dynamics of the Bacillus subtilis response regulator, Spo0F: implications for phosphorylation and molecular recognition.
Biochemistry, 36, 1997
|
|
7PSY
 
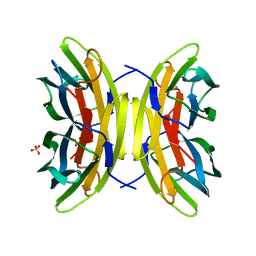 | | X-ray crystal structure of perdeuterated LecB lectin in complex with perdeuterated fucose | | 分子名称: | CALCIUM ION, Fucose-binding lectin, SULFATE ION, ... | | 著者 | Gajdos, L, Blakeley, M.P, Haertlein, M, Forsyth, T.V, Devos, J.M, Imberty, A. | | 登録日 | 2021-09-24 | | 公開日 | 2022-01-12 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (0.9 Å) | | 主引用文献 | Neutron crystallography reveals mechanisms used by Pseudomonas aeruginosa for host-cell binding.
Nat Commun, 13, 2022
|
|
7GZX
 
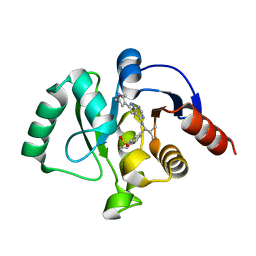 | | Crystal structure of SARS-CoV-2 NSP3 Macrodomain in complex with ASAP-0011176-001 | | 分子名称: | 7-[(1S)-2-methyl-1-{[(6M)-6-{5-[(methylamino)methyl]furan-3-yl}-7H-pyrrolo[2,3-d]pyrimidin-4-yl]amino}propyl]-3,4-dihydro-1lambda~6~-benzothiopyran-1,1(2H)-dione, Papain-like protease nsp3 | | 著者 | Aschenbrenner, J.C, Fearon, D, Tomlinson, C.W.E, Marples, P.G, Fairhead, M, Balcomb, B.H, Chandran, A.V, Godoy, A.S, Koekemoer, L, Lithgo, R.M, Ni, X, Thompson, W, Wang, S, Wild, C, Williams, E.P, Winokan, M, Walsh, M.A, von Delft, F. | | 登録日 | 2024-01-23 | | 公開日 | 2024-05-15 | | 実験手法 | X-RAY DIFFRACTION (1.15 Å) | | 主引用文献 | Group deposition of SARS-CoV-2 NSP3 Macrodomain in complex with inhibitors from the ASAP AViDD centre
To Be Published
|
|
