2V4A
 
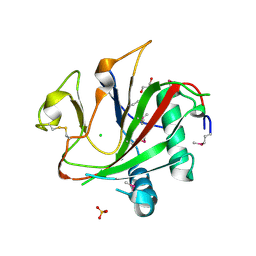 | | Crystal structure of the SeMet-labeled prolyl-4 hydroxylase (P4H) type I from green algae Chlamydomonas reinhardtii. | | 分子名称: | CHLORIDE ION, DIMETHYL SULFOXIDE, GLYCEROL, ... | | 著者 | Koski, M.K, Hieta, R, Bollner, C, Kivirikko, K.I, Myllyharju, J, Wierenga, R.K. | | 登録日 | 2007-06-28 | | 公開日 | 2007-10-30 | | 最終更新日 | 2019-07-24 | | 実験手法 | X-RAY DIFFRACTION (1.93 Å) | | 主引用文献 | The Active Site of an Algal Prolyl 4-Hydroxylase Has a Large Structural Plasticity.
J.Biol.Chem., 282, 2007
|
|
6J63
 
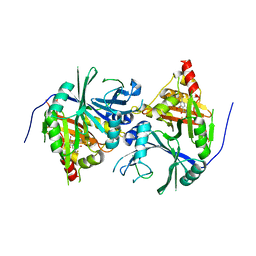 | | Crystal structure of Arabidopsis thaliana HPPD complexed with NTBC | | 分子名称: | 2-{HYDROXY[2-NITRO-4-(TRIFLUOROMETHYL)PHENYL]METHYLENE}CYCLOHEXANE-1,3-DIONE, 4-hydroxyphenylpyruvate dioxygenase, FE (III) ION | | 著者 | Yang, W.C, Yang, G.F. | | 登録日 | 2019-01-13 | | 公開日 | 2019-02-06 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.624 Å) | | 主引用文献 | Molecular insights into the mechanism of 4-hydroxyphenylpyruvate dioxygenase inhibition: enzyme kinetics, X-ray crystallography and computational simulations.
FEBS J., 286, 2019
|
|
6JB5
 
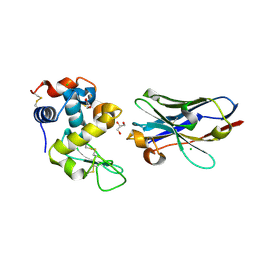 | | Crystal structure of nanobody D3-L11 mutant Y102A in complex with hen egg-white lysozyme (form II) | | 分子名称: | CHLORIDE ION, GLYCEROL, Lysozyme C, ... | | 著者 | Caaveiro, J.M.M, Tamura, H, Akiba, H, Tsumoto, K. | | 登録日 | 2019-01-25 | | 公開日 | 2019-11-06 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Structural and thermodynamic basis for the recognition of the substrate-binding cleft on hen egg lysozyme by a single-domain antibody.
Sci Rep, 9, 2019
|
|
7WUC
 
 | |
6B4O
 
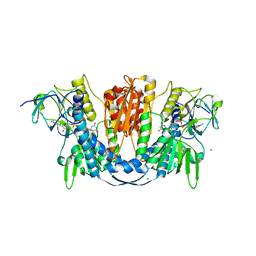 | | 1.73 Angstrom Resolution Crystal Structure of Glutathione Reductase from Enterococcus faecalis in Complex with FAD | | 分子名称: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, Glutathione reductase, ... | | 著者 | Minasov, G, Warwzak, Z, Shuvalova, L, Dubrovska, I, Cardona-Correa, A, Grimshaw, S, Kwon, K, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2017-09-27 | | 公開日 | 2017-10-11 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.73 Å) | | 主引用文献 | 1.73 Angstrom Resolution Crystal Structure of Glutathione Reductase from Enterococcus faecalis in Complex with FAD.
To Be Published
|
|
6B5I
 
 | |
5X1Y
 
 | |
2V0V
 
 | | Crystal Structure of Rev-Erb beta | | 分子名称: | ORPHAN NUCLEAR RECEPTOR NR1D2 | | 著者 | Woo, E.-J, Jeong, D.G, Lim, M.-Y, Jun Kim, S, Eon Ryu, S. | | 登録日 | 2007-05-19 | | 公開日 | 2007-10-23 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Structural Insight Into the Constitutive Repression Function of the Nuclear Receptor Rev-Erbbeta
J.Mol.Biol., 373, 2007
|
|
7WXI
 
 | | GPR domain of Drosophila P5CS filament with glutamate and ATPgammaS | | 分子名称: | Delta-1-pyrroline-5-carboxylate synthase, GAMMA-GLUTAMYL PHOSPHATE | | 著者 | Liu, J.L, Zhong, J, Guo, C.J, Zhou, X. | | 登録日 | 2022-02-14 | | 公開日 | 2022-03-30 | | 最終更新日 | 2024-06-26 | | 実験手法 | ELECTRON MICROSCOPY (4.2 Å) | | 主引用文献 | Structural basis of dynamic P5CS filaments.
Elife, 11, 2022
|
|
5X1C
 
 | |
7WXH
 
 | | GPR domain open form of Drosophila P5CS filament with glutamate, ATP, and NADPH | | 分子名称: | Delta-1-pyrroline-5-carboxylate synthase | | 著者 | Liu, J.L, Zhong, J, Guo, C.J, Zhou, X. | | 登録日 | 2022-02-14 | | 公開日 | 2022-03-30 | | 最終更新日 | 2024-06-26 | | 実験手法 | ELECTRON MICROSCOPY (4.3 Å) | | 主引用文献 | Structural basis of dynamic P5CS filaments.
Elife, 11, 2022
|
|
7WXG
 
 | | GPR domain closed form of Drosophila P5CS filament with glutamate, ATP, and NADPH | | 分子名称: | Delta-1-pyrroline-5-carboxylate synthase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | 著者 | Liu, J.L, Zhong, J, Guo, C.J, Zhou, X. | | 登録日 | 2022-02-14 | | 公開日 | 2022-03-30 | | 最終更新日 | 2024-06-26 | | 実験手法 | ELECTRON MICROSCOPY (4.2 Å) | | 主引用文献 | Structural basis of dynamic P5CS filaments.
Elife, 11, 2022
|
|
6B82
 
 | |
7WX4
 
 | |
4LRS
 
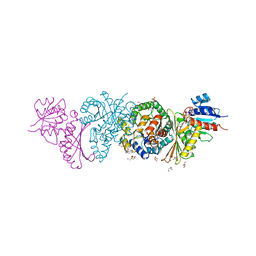 | | Crystal and solution structures of the bifunctional enzyme (Aldolase/Aldehyde dehydrogenase) from Thermomonospora curvata, reveal a cofactor-binding domain motion during NAD+ and CoA accommodation whithin the shared cofactor-binding site | | 分子名称: | 4-hydroxy-2-oxovalerate aldolase, Acetaldehyde dehydrogenase, CHLORIDE ION, ... | | 著者 | Fischer, B, Branlant, G, Talfournier, F, Gruez, A. | | 登録日 | 2013-07-20 | | 公開日 | 2013-09-04 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Crystal and solution structures of the bifunctional enzyme (Aldolase/Aldehyde dehydrogenase) from Thermomonospora curvata, reveal a cofactor-binding domain motion during NAD+ and CoA accommodation whithin the shared cofactor-binding site
To be Published
|
|
3K1O
 
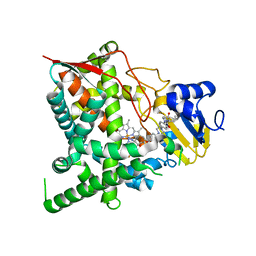 | | Crystal structure of sterol 14-alpha demethylase (CYP51) from Trypanosoma cruzi in complex with a potential antichagasic drug, posaconazole | | 分子名称: | 2,5-anhydro-1,3,4-trideoxy-2-(2,4-difluorophenyl)-6-O-{4-[4-(4-{1-[(1S,2S)-1-ethyl-2-hydroxypropyl]-5-oxo-1,5-dihydro-4H-1,2,4-triazol-4-yl}phenyl)piperazin-1-yl]phenyl}-1-(1H-1,2,4-triazol-1-yl)-D-erythro-hexitol, PROTOPORPHYRIN IX CONTAINING FE, Sterol 14 alpha-demethylase | | 著者 | Lepesheva, G.I, Hargrove, T.Y, Anderson, S, Wawrzak, Z, Waterman, M.R. | | 登録日 | 2009-09-28 | | 公開日 | 2009-10-27 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.89 Å) | | 主引用文献 | Structural Insights into Inhibition of Sterol 14{alpha}-Demethylase in the Human Pathogen Trypanosoma cruzi.
J.Biol.Chem., 285, 2010
|
|
7QHE
 
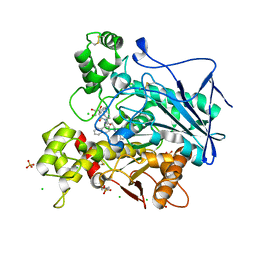 | | Human Butyrylcholinesterase in complex with (S)-1-(4-((naphthalen-1-yl)carbamoyl)benzyl)-N-(3-((1,2,3,4-tetrahydroacridin-9-yl)amino)propyl)piperidine-3-carboxamide | | 分子名称: | (3~{S})-1-[[4-(naphthalen-1-ylcarbamoyl)phenyl]methyl]-~{N}-[3-(1,2,3,4-tetrahydroacridin-9-ylamino)propyl]piperidine-3-carboxamide, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Brazzolotto, X, Jing, L, Zhan, P, Liu, X, Nachon, F. | | 登録日 | 2021-12-12 | | 公開日 | 2022-12-21 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.47 Å) | | 主引用文献 | Rapid discovery and crystallography study of highly potent and selective butylcholinesterase inhibitors based on oxime-containing libraries and conformational restriction strategies.
Bioorg.Chem., 134, 2023
|
|
2VDC
 
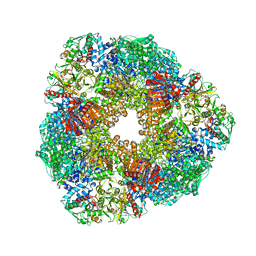 | | THE 9.5 A RESOLUTION STRUCTURE OF GLUTAMATE SYNTHASE FROM CRYO-ELECTRON MICROSCOPY AND ITS OLIGOMERIZATION BEHAVIOR IN SOLUTION: FUNCTIONAL IMPLICATIONS. | | 分子名称: | 2-OXOGLUTARIC ACID, FE3-S4 CLUSTER, FLAVIN MONONUCLEOTIDE, ... | | 著者 | Cottevieille, M, Larquet, E, Jonic, S, Petoukhov, M.V, Caprini, G, Paravisi, S, Svergun, D.I, Vanoni, M.A, Boisset, N. | | 登録日 | 2007-10-04 | | 公開日 | 2008-01-15 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (9.5 Å) | | 主引用文献 | The Subnanometer Resolution Structure of the Glutamate Synthase 1.2-Mda Hexamer by Cryoelectron Microscopy and its Oligomerization Behavior in Solution: Functional Implications.
J.Biol.Chem., 283, 2008
|
|
6FKX
 
 | | Crystal structure of an acetyl xylan esterase from a desert metagenome | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Acetyl xylan esterase, FORMIC ACID, ... | | 著者 | Adesioye, F.A, Makhalanyane, T.P, Vikram, S, Sewell, B.T, Schubert, W, Cowan, D.A. | | 登録日 | 2018-01-24 | | 公開日 | 2018-02-28 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.03 Å) | | 主引用文献 | Structural Characterization and Directed Evolution of a Novel Acetyl Xylan Esterase Reveals Thermostability Determinants of the Carbohydrate Esterase 7 Family.
Appl. Environ. Microbiol., 84, 2018
|
|
7WXF
 
 | |
6JG0
 
 | | Crystal structure of the N-terminal domain single mutant (S92E) of the human mitochondrial calcium uniporter fused with T4 lysozyme | | 分子名称: | Endolysin,Calcium uniporter protein, SULFATE ION | | 著者 | Lee, Y, Park, J, Min, C.K, Kang, J.Y, Kim, T.G, Yamamoto, T, Kim, D.H, Eom, S.H. | | 登録日 | 2019-02-13 | | 公開日 | 2020-02-19 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Crystal structure of calcium channel domain
To Be Published
|
|
3CL8
 
 | | Crystal structure of Puue Allantoinase complexed with ACA | | 分子名称: | 5-amino-1H-imidazole-4-carboxamide, Puue Allantoinase | | 著者 | Ramazzina, I, Cendron, L, Folli, C, Berni, R, Monteverdi, D, Zanotti, G, Percudani, R. | | 登録日 | 2008-03-18 | | 公開日 | 2008-06-10 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Logical identification of an allantoinase analog (puuE) recruited from polysaccharide deacetylases
J.Biol.Chem., 283, 2008
|
|
2VFR
 
 | |
7QHD
 
 | | Human Butyrylcholinesterase in complex with (S)-1-(4-((2-(1H-indol-3-yl)ethyl)carbamoyl)benzyl)-N-(3-((1,2,3,4-tetrahydroacridin-9-yl)amino)propyl)piperidine-3-carboxamide | | 分子名称: | (3~{S})-1-[[4-[2-(1~{H}-indol-3-yl)ethylcarbamoyl]phenyl]methyl]-~{N}-[3-(1,2,3,4-tetrahydroacridin-9-ylamino)propyl]piperidine-3-carboxamide, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Brazzolotto, X, Jing, L, Zhan, P, Liu, X, Nachon, F. | | 登録日 | 2021-12-12 | | 公開日 | 2022-12-21 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.04 Å) | | 主引用文献 | Rapid discovery and crystallography study of highly potent and selective butylcholinesterase inhibitors based on oxime-containing libraries and conformational restriction strategies.
Bioorg.Chem., 134, 2023
|
|
6JGN
 
 | | Crystal structure of barley exohydrolaseI W434H in complex with 4'-nitrophenyl thiolaminaribioside | | 分子名称: | 4'-NITROPHENYL-S-(BETA-D-GLUCOPYRANOSYL)-(1-3)-(3-THIO-BETA-D-GLUCOPYRANOSYL)-(1-3)-BETA-D-GLUCOPYRANOSIDE, ACETATE ION, BETA-D-GLUCAN GLUCOHYDROLASE ISOENZYME EXO1, ... | | 著者 | Luang, S, Streltsov, V.A, Hrmova, M. | | 登録日 | 2019-02-14 | | 公開日 | 2020-08-19 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.98 Å) | | 主引用文献 | The evolutionary advantage of an aromatic clamp in plant family 3 glycoside exo-hydrolases.
Nat Commun, 13, 2022
|
|
