8X15
 
 | | Structure of nucleosome-bound SRCAP-C in the apo state | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Actin, ... | | 著者 | Yu, J, Wang, Q, Yu, Z, Li, W, Wang, L, Xu, Y. | | 登録日 | 2023-11-06 | | 公開日 | 2024-03-06 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Structural insights into histone exchange by human SRCAP complex.
Cell Discov, 10, 2024
|
|
8X1C
 
 | | Structure of nucleosome-bound SRCAP-C in the ADP-bound state | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Actin, ... | | 著者 | Yu, J, Wang, Q, Yu, Z, Li, W, Wang, L, Xu, Y. | | 登録日 | 2023-11-06 | | 公開日 | 2024-03-06 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Structural insights into histone exchange by human SRCAP complex.
Cell Discov, 10, 2024
|
|
1UWB
 
 | | TYR 181 CYS HIV-1 RT/8-CL TIBO | | 分子名称: | 5-CHLORO-8-METHYL-7-(3-METHYL-BUT-2-ENYL)-6,7,8,9-TETRAHYDRO-2H-2,7,9A-TRIAZA-BENZO[CD]AZULENE-1-THIONE, REVERSE TRANSCRIPTASE | | 著者 | Das, K, Ding, J, Hsiou, Y, Arnold, E. | | 登録日 | 1996-11-21 | | 公開日 | 1997-05-15 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Crystal structures of 8-Cl and 9-Cl TIBO complexed with wild-type HIV-1 RT and 8-Cl TIBO complexed with the Tyr181Cys HIV-1 RT drug-resistant mutant.
J.Mol.Biol., 264, 1996
|
|
3E48
 
 | | Crystal structure of a nucleoside-diphosphate-sugar epimerase (SAV0421) from Staphylococcus aureus, Northeast Structural Genomics Consortium Target ZR319 | | 分子名称: | MAGNESIUM ION, Putative nucleoside-diphosphate-sugar epimerase | | 著者 | Forouhar, F, Abashidze, M, Seetharaman, J, Mao, L, Janjua, H, Xiao, R, Ciccosanti, C, Foote, E.L, Wang, D, Tong, S, Everett, J.K, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2008-08-11 | | 公開日 | 2008-08-19 | | 最終更新日 | 2025-03-26 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 |
|
|
1FKO
 
 | | CRYSTAL STRUCTURE OF NNRTI RESISTANT K103N MUTANT HIV-1 REVERSE TRANSCRIPTASE IN COMPLEX WITH DMP-266(EFAVIRENZ) | | 分子名称: | (-)-6-CHLORO-4-CYCLOPROPYLETHYNYL-4-TRIFLUOROMETHYL-1,4-DIHYDRO-2H-3,1-BENZOXAZIN-2-ONE, HIV-1 RT, A-CHAIN, ... | | 著者 | Ren, J, Milton, J, Weaver, K.L, Short, S.A, Stuart, D.I, Stammers, D.K. | | 登録日 | 2000-08-10 | | 公開日 | 2000-11-03 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Structural basis for the resilience of efavirenz (DMP-266) to drug resistance mutations in HIV-1 reverse transcriptase.
Structure Fold.Des., 8, 2000
|
|
1FK9
 
 | | CRYSTAL STRUCTURE OF HIV-1 REVERSE TRANSCRIPTASE IN COMPLEX WITH DMP-266(EFAVIRENZ) | | 分子名称: | (-)-6-CHLORO-4-CYCLOPROPYLETHYNYL-4-TRIFLUOROMETHYL-1,4-DIHYDRO-2H-3,1-BENZOXAZIN-2-ONE, HIV-1 RT, A-CHAIN, ... | | 著者 | Ren, J, Milton, J, Weaver, K.L, Short, S.A, Stuart, D.I, Stammers, D.K. | | 登録日 | 2000-08-09 | | 公開日 | 2000-11-03 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structural basis for the resilience of efavirenz (DMP-266) to drug resistance mutations in HIV-1 reverse transcriptase.
STRUCTURE FOLD.DES., 8, 2000
|
|
2WHO
 
 | |
5KMA
 
 | | Human Histidine Triad Nucleotide Binding Protein 1 (hHint1) H112N mutant nucleoside D-Trp phosphoramidate substrate complex | | 分子名称: | CHLORIDE ION, Histidine triad nucleotide-binding protein 1, [(2~{R},3~{S},4~{R},5~{R})-5-(2-azanyl-6-oxidanylidene-3~{H}-purin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-~{N}-ethyl-phosphonamidic acid | | 著者 | Maize, K.M, Finzel, B.C. | | 登録日 | 2016-06-26 | | 公開日 | 2017-06-28 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | A Crystal Structure Based Guide to the Design of Human Histidine Triad Nucleotide Binding Protein 1 (hHint1) Activated ProTides.
Mol. Pharm., 14, 2017
|
|
5KMB
 
 | | Human Histidine Triad Nucleotide Binding Protein 1 (hHint1) H112N mutant nucleoside L-Trp phosphoramidate substrate complex | | 分子名称: | CHLORIDE ION, Histidine triad nucleotide-binding protein 1, [(2~{R},3~{S},4~{R},5~{R})-5-(2-azanyl-6-oxidanylidene-1~{H}-purin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-~{N}-[(2~ {S})-3-(1~{H}-indol-3-yl)-1-(methylamino)-1-oxidanylidene-propan-2-yl]phosphonamidic acid | | 著者 | Maize, K.M, Finzel, B.C. | | 登録日 | 2016-06-26 | | 公開日 | 2017-06-28 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | A Crystal Structure Based Guide to the Design of Human Histidine Triad Nucleotide Binding Protein 1 (hHint1) Activated ProTides.
Mol. Pharm., 14, 2017
|
|
5KM6
 
 | | Human Histidine Triad Nucleotide Binding Protein 1 (hHint1) H112N mutant Ara-A nucleoside phosphoramidate substrate complex | | 分子名称: | Histidine triad nucleotide-binding protein 1, [(2~{R},3~{S},4~{S},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-~{N}-[2-(1~{H}-indol-3-yl)ethyl]phosphonamidic acid | | 著者 | Maize, K.M, Finzel, B.C. | | 登録日 | 2016-06-26 | | 公開日 | 2017-06-28 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | A Crystal Structure Based Guide to the Design of Human Histidine Triad Nucleotide Binding Protein 1 (hHint1) Activated ProTides.
Mol. Pharm., 14, 2017
|
|
5KLY
 
 | |
7EVN
 
 | | The cryo-EM structure of the DDX42-SF3b complex | | 分子名称: | ATP-dependent RNA helicase DDX42, PHD finger-like domain-containing protein 5A, Splicing factor 3B subunit 1, ... | | 著者 | Zhang, X, Zhan, X, Shi, Y. | | 登録日 | 2021-05-21 | | 公開日 | 2022-08-03 | | 最終更新日 | 2025-06-18 | | 実験手法 | ELECTRON MICROSCOPY (2.6 Å) | | 主引用文献 | Structural insights into branch site proofreading by human spliceosome
Nat.Struct.Mol.Biol., 2024
|
|
2RKI
 
 | | Crystal Structure of HIV-1 Reverse Transcriptase (RT) in Complex with a triazole derived NNRTI | | 分子名称: | 4-benzyl-3-[(2-chlorobenzyl)sulfanyl]-5-thiophen-2-yl-4H-1,2,4-triazole, CHLORIDE ION, GLYCEROL, ... | | 著者 | Lansdon, E.B, Kirschberg, T.A. | | 登録日 | 2007-10-16 | | 公開日 | 2008-04-22 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Triazole derivatives as non-nucleoside inhibitors of HIV-1 reverse transcriptase-structure-activity relationships and crystallographic analysis.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
1C1B
 
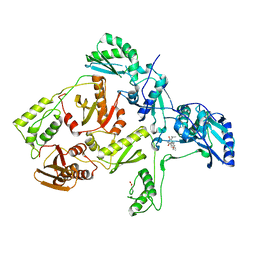 | | CRYSTAL STRUCTURE OF HIV-1 REVERSE TRANSCRIPTASE IN COMPLEX WITH GCA-186 | | 分子名称: | 6-(3',5'-DIMETHYLBENZYL)-1-ETHOXYMETHYL-5-ISOPROPYLURACIL, HIV-1 REVERSE TRANSCRIPTASE (A-CHAIN), HIV-1 REVERSE TRANSCRIPTASE (B-CHAIN) | | 著者 | Hopkins, A.L, Ren, J, Tanaka, H, Baba, B, Okamato, M, Stuart, D.I, Stammers, D.K. | | 登録日 | 1999-07-21 | | 公開日 | 2000-07-21 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Design of MKC-442 (emivirine) analogues with improved activity against drug-resistant HIV mutants.
J.Med.Chem., 42, 1999
|
|
1C1C
 
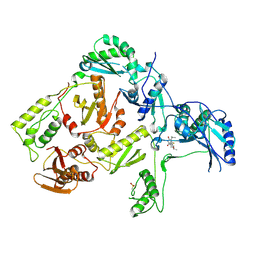 | | CRYSTAL STRUCTURE OF HIV-1 REVERSE TRANSCRIPTASE IN COMPLEX WITH TNK-6123 | | 分子名称: | 6-(cyclohexylsulfanyl)-1-(ethoxymethyl)-5-(1-methylethyl)pyrimidine-2,4(1H,3H)-dione, HIV-1 REVERSE TRANSCRIPTASE (A-CHAIN), HIV-1 REVERSE TRANSCRIPTASE (B-CHAIN) | | 著者 | Hopkins, A.L, Ren, J, Tanaka, H, Baba, M, Okamato, M, Stuart, D.I, Stammers, D.K. | | 登録日 | 1999-07-21 | | 公開日 | 2000-07-21 | | 最終更新日 | 2024-11-20 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Design of MKC-442 (emivirine) analogues with improved activity against drug-resistant HIV mutants.
J.Med.Chem., 42, 1999
|
|
3K8T
 
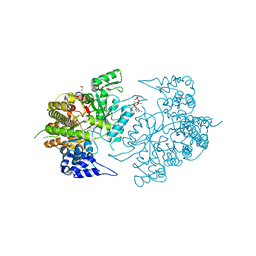 | | Structure of eukaryotic rnr large subunit R1 complexed with designed adp analog compound | | 分子名称: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, 2'-deoxy-2'-(2-hydroxyethyl)adenosine 5'-(trihydrogen diphosphate), GLYCEROL, ... | | 著者 | Sun, D, Xu, H, Dealwis, C, Lee, R.E. | | 登録日 | 2009-10-14 | | 公開日 | 2010-03-02 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structure-Based Design, Synthesis, and Evaluation of 2'-(2-Hydroxyethyl)-2'-deoxyadenosine and the 5'-Diphosphate Derivative as Ribonucleotide Reductase Inhibitors
Chemmedchem, 4, 2009
|
|
7O80
 
 | | Rabbit 80S ribosome in complex with eRF1 and ABCE1 stalled at the STOP codon in the mutated SARS-CoV-2 slippery site | | 分子名称: | 18S rRNA, 28S rRNA, 40S ribosomal protein S11, ... | | 著者 | Bhatt, P.R, Scaiola, A, Leibundgut, M.A, Atkins, J.F, Ban, N. | | 登録日 | 2021-04-14 | | 公開日 | 2021-06-02 | | 最終更新日 | 2024-04-24 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Structural basis of ribosomal frameshifting during translation of the SARS-CoV-2 RNA genome.
Science, 372, 2021
|
|
7O81
 
 | | Rabbit 80S ribosome colliding in another ribosome stalled by the SARS-CoV-2 pseudoknot | | 分子名称: | 18S rRNA, 28S rRNA, 40S ribosomal protein S11, ... | | 著者 | Bhatt, P.R, Scaiola, A, Leibundgut, M.A, Atkins, J.F, Ban, N. | | 登録日 | 2021-04-14 | | 公開日 | 2021-06-02 | | 最終更新日 | 2024-04-24 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Structural basis of ribosomal frameshifting during translation of the SARS-CoV-2 RNA genome.
Science, 372, 2021
|
|
7O7Z
 
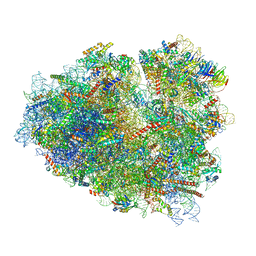 | | Rabbit 80S ribosome stalled close to the mutated SARS-CoV-2 slippery site by a pseudoknot (classified for pseudoknot) | | 分子名称: | 18S rRNA, 28S rRNA, 40S ribosomal protein S11, ... | | 著者 | Bhatt, P.R, Scaiola, A, Leibundgut, M.A, Atkins, J.F, Ban, N. | | 登録日 | 2021-04-14 | | 公開日 | 2021-06-02 | | 最終更新日 | 2024-04-24 | | 実験手法 | ELECTRON MICROSCOPY (2.4 Å) | | 主引用文献 | Structural basis of ribosomal frameshifting during translation of the SARS-CoV-2 RNA genome.
Science, 372, 2021
|
|
7O7Y
 
 | | Rabbit 80S ribosome stalled close to the mutated SARS-CoV-2 slippery site by a pseudoknot (high resolution) | | 分子名称: | 18S rRNA, 28S rRNA, 40S ribosomal protein S11, ... | | 著者 | Bhatt, P.R, Scaiola, A, Leibundgut, M.A, Atkins, J.F, Ban, N. | | 登録日 | 2021-04-14 | | 公開日 | 2021-06-02 | | 最終更新日 | 2024-04-24 | | 実験手法 | ELECTRON MICROSCOPY (2.2 Å) | | 主引用文献 | Structural basis of ribosomal frameshifting during translation of the SARS-CoV-2 RNA genome.
Science, 372, 2021
|
|
1U35
 
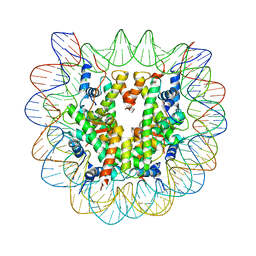 | | Crystal structure of the nucleosome core particle containing the histone domain of macroH2A | | 分子名称: | H2A histone family, Hist1h4i protein, Histone H3.1, ... | | 著者 | Chakravarthy, S, Gundimella, S.K, Caron, C, Perche, P.Y, Pehrson, J.R, Khochbin, S, Luger, K. | | 登録日 | 2004-07-20 | | 公開日 | 2005-09-27 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Structural characterization of the histone variant macroH2A.
Mol.Cell.Biol., 25, 2005
|
|
5TER
 
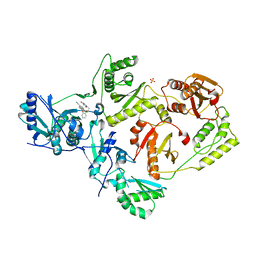 | | Crystal Structure of HIV-1 Reverse Transcriptase in Complex with 5-chloro-7-(2-(2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy)phenoxy)-8-methyl-2-naphthonitrile (JLJ651), a Non-nucleoside Inhibitor | | 分子名称: | 5-chloro-7-(2-(2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy)phenoxy)-8-methyl-2-naphthonitrile, HIV-1 REVERSE TRANSCRIPTASE, P51 SUBUNIT, ... | | 著者 | Chan, A.H, Anderson, K.S. | | 登録日 | 2016-09-22 | | 公開日 | 2017-01-11 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Design, Conformation, and Crystallography of 2-Naphthyl Phenyl Ethers as Potent Anti-HIV Agents.
ACS Med Chem Lett, 7, 2016
|
|
1TVR
 
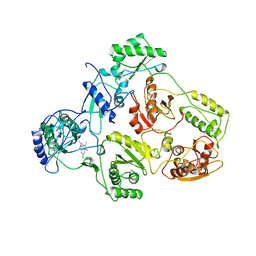 | | HIV-1 RT/9-CL TIBO | | 分子名称: | 4-CHLORO-8-METHYL-7-(3-METHYL-BUT-2-ENYL)-6,7,8,9-TETRAHYDRO-2H-2,7,9A-TRIAZA-BENZO[CD]AZULENE-1-THIONE, REVERSE TRANSCRIPTASE | | 著者 | Das, K, Ding, J, Hsiou, Y, Arnold, E. | | 登録日 | 1996-04-16 | | 公開日 | 1997-03-12 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Crystal structures of 8-Cl and 9-Cl TIBO complexed with wild-type HIV-1 RT and 8-Cl TIBO complexed with the Tyr181Cys HIV-1 RT drug-resistant mutant.
J.Mol.Biol., 264, 1996
|
|
4G1Q
 
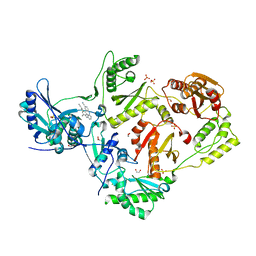 | | Crystal structure of HIV-1 reverse transcriptase (RT) in complex with Rilpivirine (TMC278, Edurant), a non-nucleoside rt-inhibiting drug | | 分子名称: | 1,2-ETHANEDIOL, 4-{[4-({4-[(E)-2-cyanoethenyl]-2,6-dimethylphenyl}amino)pyrimidin-2-yl]amino}benzonitrile, MAGNESIUM ION, ... | | 著者 | Bauman, J.D, Patel, D, Das, K, Arnold, E. | | 登録日 | 2012-07-11 | | 公開日 | 2013-02-06 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.51 Å) | | 主引用文献 | Snapshot of the equilibrium dynamics of a drug bound to HIV-1 reverse transcriptase.
Nat Chem, 5, 2013
|
|
1VRU
 
 | | HIGH RESOLUTION STRUCTURES OF HIV-1 RT FROM FOUR RT-INHIBITOR COMPLEXES | | 分子名称: | ALPHA-(2,6-DICHLOROPHENYL)-ALPHA-(2-ACETYL-5-METHYLANILINO)ACETAMIDE, HIV-1 REVERSE TRANSCRIPTASE | | 著者 | Ren, J, Esnouf, R, Garman, E, Somers, D, Ross, C, Kirby, I, Keeling, J, Darby, G, Jones, Y, Stuart, D, Stammers, D. | | 登録日 | 1995-04-19 | | 公開日 | 1996-04-03 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | High resolution structures of HIV-1 RT from four RT-inhibitor complexes.
Nat.Struct.Biol., 2, 1995
|
|
