4G8C
 
 | |
4G8D
 
 | |
4G9E
 
 | |
4G9G
 
 | |
4GW3
 
 | | Crystal Structure of the Lipase from Proteus mirabilis | | 分子名称: | CALCIUM ION, GLYCEROL, ISOPROPYL ALCOHOL, ... | | 著者 | Korman, T.P. | | 登録日 | 2012-08-31 | | 公開日 | 2013-02-06 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal Structure of Proteus mirabilis Lipase, a Novel Lipase from the Proteus/Psychrophilic Subfamily of Lipase Family I.1.
Plos One, 7, 2012
|
|
4GXN
 
 | |
4H77
 
 | | Crystal structure of haloalkane dehalogenase LinB from Sphingobium sp. MI1205 | | 分子名称: | CALCIUM ION, CHLORIDE ION, GLYCEROL, ... | | 著者 | Okai, M, Ohtsuka, J, Imai, F.L, Mase, T, Moriuchi, R, Tsuda, M, Nagata, K, Nagata, Y, Tanokura, M. | | 登録日 | 2012-09-20 | | 公開日 | 2013-06-05 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Crystal Structure and Site-Directed Mutagenesis Analyses of Haloalkane Dehalogenase LinB from Sphingobium sp. Strain MI1205.
J.Bacteriol., 195, 2013
|
|
4H7D
 
 | | Crystal structure of haloalkane dehalogenase LinB T81A mutant from Sphingobium sp. MI1205 | | 分子名称: | CALCIUM ION, CHLORIDE ION, GLYCEROL, ... | | 著者 | Okai, M, Ohtsuka, J, Imai, L.F, Mase, T, Moriuchi, R, Tsuda, M, Nagata, K, Nagata, Y, Tanokura, M. | | 登録日 | 2012-09-20 | | 公開日 | 2013-06-05 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Crystal Structure and Site-Directed Mutagenesis Analyses of Haloalkane Dehalogenase LinB from Sphingobium sp. Strain MI1205.
J.Bacteriol., 195, 2013
|
|
4H7E
 
 | | Crystal structure of haloalkane dehalogenase LinB V112A mutant from Sphingobium sp. MI1205 | | 分子名称: | CALCIUM ION, CHLORIDE ION, GLYCEROL, ... | | 著者 | Okai, M, Ohtsuka, J, Imai, L.F, Mase, T, Moriuchi, R, Tsuda, M, Nagata, K, Nagata, Y, Tanokura, M. | | 登録日 | 2012-09-20 | | 公開日 | 2013-06-05 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal Structure and Site-Directed Mutagenesis Analyses of Haloalkane Dehalogenase LinB from Sphingobium sp. Strain MI1205.
J.Bacteriol., 195, 2013
|
|
4H7F
 
 | | Crystal structure of haloalkane dehalogenase LinB V134I mutant from Sphingobium sp. MI1205 | | 分子名称: | CALCIUM ION, CHLORIDE ION, GLYCEROL, ... | | 著者 | Okai, M, Ohtsuka, J, Imai, L.F, Mase, T, Moriuchi, R, Tsuda, M, Nagata, K, Nagata, Y, Tanokura, M. | | 登録日 | 2012-09-20 | | 公開日 | 2013-06-05 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal Structure and Site-Directed Mutagenesis Analyses of Haloalkane Dehalogenase LinB from Sphingobium sp. Strain MI1205.
J.Bacteriol., 195, 2013
|
|
4H7H
 
 | | Crystal structure of haloalkane dehalogenase LinB T135A mutant from Sphingobium sp. MI1205 | | 分子名称: | CALCIUM ION, CHLORIDE ION, Haloalkane dehalogenase | | 著者 | Okai, M, Ohtsuka, J, Imai, L.F, Mase, T, Moriuchi, R, Tsuda, M, Nagata, K, Nagata, Y, Tanokura, M. | | 登録日 | 2012-09-20 | | 公開日 | 2013-06-05 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Crystal Structure and Site-Directed Mutagenesis Analyses of Haloalkane Dehalogenase LinB from Sphingobium sp. Strain MI1205.
J.Bacteriol., 195, 2013
|
|
4H7I
 
 | | Crystal structure of haloalkane dehalogenase LinB L138I mutant from Sphingobium sp. MI1205 | | 分子名称: | CALCIUM ION, CHLORIDE ION, GLYCEROL, ... | | 著者 | Okai, M, Ohtsuka, J, Imai, L.F, Mase, T, Moriuchi, R, Tsuda, M, Nagata, K, Nagata, Y, Tanokura, M. | | 登録日 | 2012-09-20 | | 公開日 | 2013-06-05 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal Structure and Site-Directed Mutagenesis Analyses of Haloalkane Dehalogenase LinB from Sphingobium sp. Strain MI1205.
J.Bacteriol., 195, 2013
|
|
4H7J
 
 | | Crystal structure of haloalkane dehalogenase LinB H247A mutant from Sphingobium sp. MI1205 | | 分子名称: | CALCIUM ION, CHLORIDE ION, GLYCEROL, ... | | 著者 | Okai, M, Ohtsuka, J, Imai, L.F, Mase, T, Moriuchi, R, Tsuda, M, Nagata, K, Nagata, Y, Tanokura, M. | | 登録日 | 2012-09-20 | | 公開日 | 2013-06-05 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal Structure and Site-Directed Mutagenesis Analyses of Haloalkane Dehalogenase LinB from Sphingobium sp. Strain MI1205.
J.Bacteriol., 195, 2013
|
|
4H7K
 
 | | Crystal structure of haloalkane dehalogenase LinB I253M mutant from Sphingobium sp. MI1205 | | 分子名称: | CALCIUM ION, CHLORIDE ION, Haloalkane dehalogenase | | 著者 | Okai, M, Ohtsuka, J, Imai, L.F, Mase, T, Moriuchi, R, Tsuda, M, Nagata, K, Nagata, Y, Tanokura, M. | | 登録日 | 2012-09-20 | | 公開日 | 2013-06-05 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Crystal Structure and Site-Directed Mutagenesis Analyses of Haloalkane Dehalogenase LinB from Sphingobium sp. Strain MI1205.
J.Bacteriol., 195, 2013
|
|
4HAI
 
 | | Crystal structure of human soluble epoxide hydrolase complexed with N-cycloheptyl-1-(mesitylsulfonyl)piperidine-4-carboxamide. | | 分子名称: | Bifunctional epoxide hydrolase 2, MAGNESIUM ION, N-cycloheptyl-1-[(2,4,6-trimethylphenyl)sulfonyl]piperidine-4-carboxamide, ... | | 著者 | Pecic, S, Pakhomova, S, Newcomer, M.E, Morisseau, C, Hammock, B.D, Zhu, Z, Deng, S. | | 登録日 | 2012-09-26 | | 公開日 | 2012-12-26 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.55 Å) | | 主引用文献 | Synthesis and structure-activity relationship of piperidine-derived non-urea soluble epoxide hydrolase inhibitors.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
4HS9
 
 | | Methanol tolerant mutant of the Proteus mirabilis lipase | | 分子名称: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, CALCIUM ION, CHLORIDE ION, ... | | 著者 | Korman, T.P. | | 登録日 | 2012-10-29 | | 公開日 | 2013-05-22 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Dieselzymes: development of a stable and methanol tolerant lipase for biodiesel production by directed evolution.
Biotechnol Biofuels, 6, 2013
|
|
4HZG
 
 | | Structure of haloalkane dehalogenase DhaA from Rhodococcus rhodochrous | | 分子名称: | CHLORIDE ION, Haloalkane dehalogenase | | 著者 | Stsiapanava, A, Weiss, M.S, Mesters, J.R, Kuta Smatanova, I. | | 登録日 | 2012-11-15 | | 公開日 | 2013-10-02 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Crystallographic analysis of 1,2,3-trichloropropane biodegradation by the haloalkane dehalogenase DhaA31.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4I3F
 
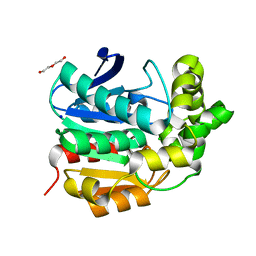 | | Crystal structure of serine hydrolase CCSP0084 from the polyaromatic hydrocarbon (PAH)-degrading bacterium Cycloclasticus zankles | | 分子名称: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, SODIUM ION, ... | | 著者 | Stogios, P.J, Xu, X, Dong, A, Cui, H, Alcaide, M, Tornes, J, Gertler, C, Yakimov, M.M, Golyshin, P.N, Ferrer, M, Savchenko, A. | | 登録日 | 2012-11-26 | | 公開日 | 2013-06-26 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.69 Å) | | 主引用文献 | Single residues dictate the co-evolution of dual esterases: MCP hydrolases from the alpha / beta hydrolase family.
Biochem.J., 454, 2013
|
|
4INZ
 
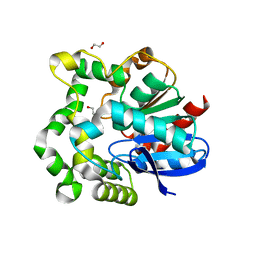 | | The crystal structure of M145A mutant of an epoxide hydrolase from Bacillus megaterium | | 分子名称: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Soluble epoxide hydrolase | | 著者 | Kong, X.D, Zhou, J.H, Xu, J.H. | | 登録日 | 2013-01-07 | | 公開日 | 2014-02-12 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Engineering of an epoxide hydrolase for efficient bioresolution of bulky pharmaco substrates.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4IO0
 
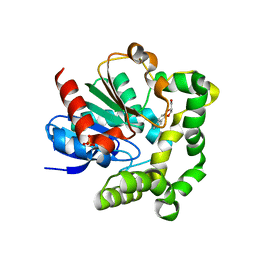 | | Crystal structure of F128A mutant of an epoxide hydrolase from Bacillus megaterium complexed with its product (R)-3-[1]naphthyloxy-propane-1,2-diol | | 分子名称: | (2R)-3-(naphthalen-1-yloxy)propane-1,2-diol, SULFATE ION, Soluble epoxide hydrolase | | 著者 | Kong, X.D, Zhou, J.H, Xu, J.H. | | 登録日 | 2013-01-07 | | 公開日 | 2014-02-12 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Engineering of an epoxide hydrolase for efficient bioresolution of bulky pharmaco substrates.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4IQ4
 
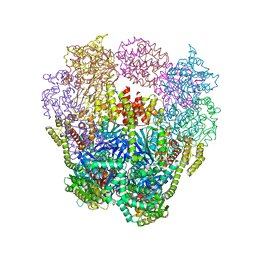 | | Structure of a 16 nm protein cage designed by fusing symmetric oligomeric domains, triple mutant, P21212 form | | 分子名称: | Non-haem bromoperoxidase BPO-A2, Matrix protein 1 | | 著者 | Lai, Y.-T, Sawaya, M.R, Yeates, T.O. | | 登録日 | 2013-01-10 | | 公開日 | 2013-07-24 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (3.495 Å) | | 主引用文献 | Structure and flexibility of nanoscale protein cages designed by symmetric self-assembly.
J.Am.Chem.Soc., 135, 2013
|
|
4ITV
 
 | | Structure of a 16 nm protein cage designed by fusing symmetric oligomeric domains, triple mutant, P212121 form | | 分子名称: | Non-haem bromoperoxidase BPO-A2, Matrix protein 1 | | 著者 | Lai, Y.-T, Sawaya, M.R, Yeates, T.O. | | 登録日 | 2013-01-18 | | 公開日 | 2013-07-24 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (3.598 Å) | | 主引用文献 | Structure and flexibility of nanoscale protein cages designed by symmetric self-assembly.
J.Am.Chem.Soc., 135, 2013
|
|
4IVJ
 
 | | Structure of a 16 nm protein cage designed by fusing symmetric oligomeric domains, triple mutant, I222 form | | 分子名称: | Non-haem bromoperoxidase BPO-A2, Matrix protein 1 | | 著者 | Lai, Y.-T, Sawaya, M.R, Yeates, T.O. | | 登録日 | 2013-01-23 | | 公開日 | 2013-07-24 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (7.351 Å) | | 主引用文献 | Structure and flexibility of nanoscale protein cages designed by symmetric self-assembly.
J.Am.Chem.Soc., 135, 2013
|
|
4J03
 
 | | Crystal structure of human soluble epoxide hydrolase complexed with fulvestrant | | 分子名称: | (7beta,9beta,13alpha,17beta)-7-{9-[(R)-(4,4,5,5,5-pentafluoropentyl)sulfinyl]nonyl}estra-1(10),2,4-triene-3,17-diol, Bifunctional epoxide hydrolase 2, MAGNESIUM ION, ... | | 著者 | Morisseau, C, Pakhomova, S, Hwang, S.H, Newcomer, M.E, Hammock, B.D. | | 登録日 | 2013-01-30 | | 公開日 | 2013-06-05 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.92 Å) | | 主引用文献 | Inhibition of soluble epoxide hydrolase by fulvestrant and sulfoxides.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
4JNC
 
 | | Soluble Epoxide Hydrolase complexed with a carboxamide inhibitor | | 分子名称: | 1-[4-methyl-6-(methylamino)-1,3,5-triazin-2-yl]-N-[2-(trifluoromethyl)benzyl]piperidine-4-carboxamide, Bifunctional epoxide hydrolase 2 | | 著者 | Shewchuk, L.M. | | 登録日 | 2013-03-15 | | 公開日 | 2013-06-05 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.96 Å) | | 主引用文献 | Discovery of 1-(1,3,5-triazin-2-yl)piperidine-4-carboxamides as inhibitors of soluble epoxide hydrolase.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
