4RME
 
 | | Crystal structure of human Retinoid X receptor alpha ligand binding domain complex with 9cUAB111 and coactivator peptide GRIP-1 | | 分子名称: | (2E,4E,6Z,8E)-3,7-dimethyl-8-[2-(3-methylbutyl)-3-(propan-2-yl)cyclohex-2-en-1-ylidene]octa-2,4,6-trienoic acid, Nuclear receptor coactivator 2, Retinoic acid receptor RXR-alpha | | 著者 | Xia, G, Muccio, D.D. | | 登録日 | 2014-10-21 | | 公開日 | 2015-09-16 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Conformationally Defined Rexinoids and Their Efficacy in the Prevention of Mammary Cancers.
J.Med.Chem., 58, 2015
|
|
7G02
 
 | | Crystal Structure of human FABP4 in complex with 7-methoxy-1-(3-methoxyphenyl)naphthalene-2-carboxylic acid, i.e. SMILES c1(c2c(ccc1C(=O)O)ccc(c2)OC)c1cc(ccc1)OC with IC50=0.045 microM | | 分子名称: | (1M)-7-methoxy-1-(3-methoxyphenyl)naphthalene-2-carboxylic acid, 1,2-ETHANEDIOL, FORMIC ACID, ... | | 著者 | Ehler, A, Benz, J, Obst, U, Guthrie-Robert, W, Rudolph, M.G. | | 登録日 | 2023-04-27 | | 公開日 | 2023-06-14 | | 最終更新日 | 2025-08-13 | | 実験手法 | X-RAY DIFFRACTION (1.03 Å) | | 主引用文献 | A high-resolution data set of fatty acid-binding protein structures. III. Unexpectedly high occurrence of wrong ligands.
Acta Crystallogr D Struct Biol, 81, 2025
|
|
1Z9E
 
 | | Solution structure of the HIV-1 integrase-binding domain in LEDGF/p75 | | 分子名称: | PC4 and SFRS1 interacting protein 2 | | 著者 | Cherepanov, P, Sun, Z.-Y.J, Rahman, S, Maertens, G, Wagner, G, Engelman, A. | | 登録日 | 2005-04-01 | | 公開日 | 2005-05-17 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of the HIV-1 integrase-binding domain in LEDGF/p75
Nat.Struct.Mol.Biol., 12, 2005
|
|
6KK3
 
 | | Crystal structure of Zika NS2B-NS3 protease with compound 4 | | 分子名称: | 1-[(10~{R},17~{S},20~{S})-17,20-bis(4-azanylbutyl)-4,9,16,19,22-pentakis(oxidanylidene)-3,8,15,18,21-pentazabicyclo[22.2.2]octacosa-1(26),24,27-trien-10-yl]guanidine, Genome polyprotein | | 著者 | Quek, J.P. | | 登録日 | 2019-07-23 | | 公開日 | 2020-06-17 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Structure-Based Macrocyclization of Substrate Analogue NS2B-NS3 Protease Inhibitors of Zika, West Nile and Dengue viruses.
Chemmedchem, 15, 2020
|
|
3OV2
 
 | | Curcumin synthase 1 from Curcuma longa | | 分子名称: | 1,2-ETHANEDIOL, Curcumin synthase, MALONATE ION | | 著者 | Katsuyama, Y, Miyazono, K, Tanokura, M, Ohnishi, Y, Horinouchi, S. | | 登録日 | 2010-09-15 | | 公開日 | 2010-12-08 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.32 Å) | | 主引用文献 | A hydrophobic cavity discovered in a curcumin synthase facilitates utilization of a beta-keto acid as an extender substrate for the atypical type III polyleteide synthase
To be Published
|
|
3TZW
 
 | | Crystal structure of a fragment containing the acyltransferase domain of Pks13 from Mycobacterium tuberculosis in the orthorhombic apoform at 2.6 A | | 分子名称: | 1,2-ETHANEDIOL, 12-mer peptide, Polyketide synthase PKS13, ... | | 著者 | Bergeret, F, Pedelacq, J.D, Mourey, L, Bon, C. | | 登録日 | 2011-09-28 | | 公開日 | 2012-08-29 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Biochemical and structural study of the atypical acyltransferase domain from the mycobacterial polyketide synthase pks13
J.Biol.Chem., 287, 2012
|
|
1DP7
 
 | | COCRYSTAL STRUCTURE OF RFX-DBD IN COMPLEX WITH ITS COGNATE X-BOX BINDING SITE | | 分子名称: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, DNA (5'-D(*CP*GP*(BRU)P*TP*AP*CP*CP*AP*(BRU)P*GP*GP*TP*AP*AP*CP*G)-3'), ... | | 著者 | Gajiwala, K.S, Chen, H, Cornille, F, Roques, B.P, Reith, W, Mach, B, Burley, S.K. | | 登録日 | 1999-12-23 | | 公開日 | 2000-03-06 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Structure of the winged-helix protein hRFX1 reveals a new mode of DNA binding.
Nature, 403, 2000
|
|
3P0T
 
 | |
5VFC
 
 | | WDR5 bound to inhibitor MM-589 | | 分子名称: | 1,2-ETHANEDIOL, N-{(3R,6S,9S,12R)-6-ethyl-12-methyl-9-[3-(N'-methylcarbamimidamido)propyl]-2,5,8,11-tetraoxo-3-phenyl-1,4,7,10-tetraazacyclotetradecan-12-yl}-2-methylpropanamide, SULFATE ION, ... | | 著者 | Stuckey, J.A. | | 登録日 | 2017-04-07 | | 公開日 | 2017-06-28 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.64 Å) | | 主引用文献 | Discovery of a Highly Potent, Cell-Permeable Macrocyclic Peptidomimetic (MM-589) Targeting the WD Repeat Domain 5 Protein (WDR5)-Mixed Lineage Leukemia (MLL) Protein-Protein Interaction.
J. Med. Chem., 60, 2017
|
|
7FYX
 
 | | Crystal Structure of human FABP4 in complex with 2-[(3-phenylthiophen-2-yl)carbamoyl]cyclopentene-1-carboxylic acid, i.e. SMILES S1C(=C(C=C1)c1ccccc1)NC(=O)C1=C(CCC1)C(=O)O with IC50=0.201 microM | | 分子名称: | 2-[(3-phenylthiophen-2-yl)carbamoyl]cyclopent-1-ene-1-carboxylic acid, Fatty acid-binding protein, adipocyte, ... | | 著者 | Ehler, A, Benz, J, Obst, U, Ceccarelli-Simona, M, Rudolph, M.G. | | 登録日 | 2023-04-27 | | 公開日 | 2023-06-14 | | 最終更新日 | 2025-08-13 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | A high-resolution data set of fatty acid-binding protein structures. III. Unexpectedly high occurrence of wrong ligands.
Acta Crystallogr D Struct Biol, 81, 2025
|
|
7FYV
 
 | | Crystal Structure of human FABP4 in complex with 5-(4-chlorophenyl)-9-methyl-3-oxa-4-azatricyclo[5.2.1.02,6]dec-4-ene-8-carboxylic acid, i.e. SMILES [C@H]12[C@H]3[C@@H]([C@@H]([C@@H]([C@H]1ON=C2c1ccc(cc1)Cl)C3)C)C(=O)O with IC50=5.6 microM | | 分子名称: | (3aR,4R,5R,6S,7R,7aR)-3-(4-chlorophenyl)-6-methyl-3a,4,5,6,7,7a-hexahydro-4,7-methano-1,2-benzoxazole-5-carboxylic acid, 1,2-ETHANEDIOL, Fatty acid-binding protein, ... | | 著者 | Ehler, A, Benz, J, Obst, U, Wickens, J, Rudolph, M.G. | | 登録日 | 2023-04-27 | | 公開日 | 2023-06-14 | | 最終更新日 | 2025-08-13 | | 実験手法 | X-RAY DIFFRACTION (0.97 Å) | | 主引用文献 | A high-resolution data set of fatty acid-binding protein structures. III. Unexpectedly high occurrence of wrong ligands.
Acta Crystallogr D Struct Biol, 81, 2025
|
|
3E8M
 
 | | Structure-function Analysis of 2-Keto-3-deoxy-D-glycero-D-galacto-nononate-9-phosphate (KDN) Phosphatase Defines a New Clad Within the Type C0 HAD Subfamily | | 分子名称: | 1,2-ETHANEDIOL, ACETIC ACID, Acylneuraminate cytidylyltransferase, ... | | 著者 | Lu, Z, Wang, L, Dunaway-Mariano, D, Allen, K.N. | | 登録日 | 2008-08-20 | | 公開日 | 2008-11-04 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.1 Å) | | 主引用文献 | Structure-Function Analysis of 2-Keto-3-deoxy-D-glycero-D-galactonononate-9-phosphate Phosphatase Defines Specificity Elements in Type C0 Haloalkanoate Dehalogenase Family Members.
J.Biol.Chem., 284, 2009
|
|
1C9Y
 
 | | HUMAN ORNITHINE TRANSCARBAMYLASE: CRYSTALLOGRAPHIC INSIGHTS INTO SUBSTRATE RECOGNITION AND CATALYTIC MECHANISM | | 分子名称: | NORVALINE, ORNITHINE CARBAMOYLTRANSFERASE, PHOSPHORIC ACID MONO(FORMAMIDE)ESTER | | 著者 | Shi, D, Yu, X, Morizono, H, Tuchman, M, Allewell, N.M. | | 登録日 | 1999-08-03 | | 公開日 | 2000-06-06 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystal structure of human ornithine transcarbamylase complexed with carbamoyl phosphate and L-norvaline at 1.9 A resolution.
Proteins, 39, 2000
|
|
5EW9
 
 | |
1V5S
 
 | | Solution structure of kinase associated domain 1 of mouse MAP/microtubule affinity-regulating kinase 3 | | 分子名称: | MAP/microtubule affinity-regulating kinase 3 | | 著者 | Tochio, N, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2003-11-25 | | 公開日 | 2004-05-25 | | 最終更新日 | 2023-12-27 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of kinase associated domain 1 of mouse MAP/microtubule affinity-regulating kinase 3
To be Published
|
|
5LVC
 
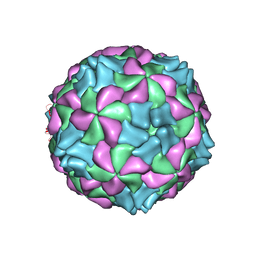 | | Aichi virus 1: empty particle | | 分子名称: | VP0, VP1, VP3 | | 著者 | Sabin, C, Fuzik, T, Skubnik, K, Palkova, L, Lindberg, A.M, Plevka, P. | | 登録日 | 2016-09-13 | | 公開日 | 2016-12-14 | | 最終更新日 | 2024-05-15 | | 実験手法 | ELECTRON MICROSCOPY (4.2 Å) | | 主引用文献 | Structure of Aichi Virus 1 and Its Empty Particle: Clues to Kobuvirus Genome Release Mechanism.
J.Virol., 90, 2016
|
|
3OXP
 
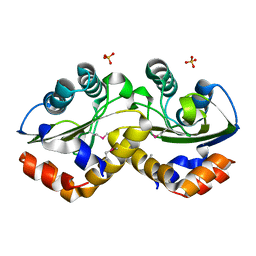 | | Structure of phosphotransferase enzyme II, A component from Yersinia pestis CO92 at 1.2 A resolution | | 分子名称: | GLYCEROL, Phosphotransferase enzyme II, A component, ... | | 著者 | Filippova, E.V, Wawrzak, Z, Kudritska, M, Edwards, A, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2010-09-21 | | 公開日 | 2010-10-13 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (1.2 Å) | | 主引用文献 | Structure of phosphotransferase enzyme II, A component from Yersinia pestis CO92 at 1.2 A resolution
To be Published
|
|
5LF4
 
 | | Human 20S proteasome complex with Delanzomib at 2.0 Angstrom | | 分子名称: | CHLORIDE ION, MAGNESIUM ION, PENTAETHYLENE GLYCOL, ... | | 著者 | Schrader, J, Henneberg, F, Mata, R, Tittmann, K, Schneider, T.R, Stark, H, Bourenkov, G, Chari, A. | | 登録日 | 2016-06-30 | | 公開日 | 2016-08-17 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.99 Å) | | 主引用文献 | The inhibition mechanism of human 20S proteasomes enables next-generation inhibitor design.
Science, 353, 2016
|
|
3ZGE
 
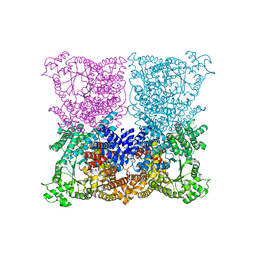 | | Greater efficiency of photosynthetic carbon fixation due to single amino acid substitution | | 分子名称: | 1,2-ETHANEDIOL, ASPARTIC ACID, C4 PHOSPHOENOLPYRUVATE CARBOXYLASE, ... | | 著者 | Paulus, J.K, Schlieper, D, Groth, G. | | 登録日 | 2012-12-17 | | 公開日 | 2013-02-27 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.49 Å) | | 主引用文献 | Greater Efficiency of Photosynthetic Carbon Fixation due to Single Amino Acid Substitution
Nat.Commun., 4, 2013
|
|
3TYP
 
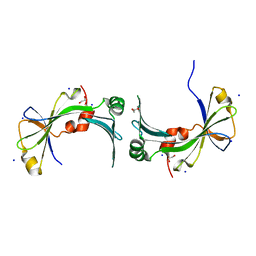 | | The crystal structure of the inorganic triphosphatase NE1496 | | 分子名称: | 1,2-ETHANEDIOL, SODIUM ION, Uncharacterized protein | | 著者 | Lunin, V.V, Skarina, T, Onopriyenko, O, Binkowski, T.A, Joachimiak, A, Edwards, A.M, Savchenko, A. | | 登録日 | 2011-09-26 | | 公開日 | 2012-05-09 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | A specific inorganic triphosphatase from Nitrosomonas europaea: structure and catalytic mechanism.
J.Biol.Chem., 286, 2011
|
|
1EPT
 
 | | REFINED 1.8 ANGSTROMS RESOLUTION CRYSTAL STRUCTURE OF PORCINE EPSILON-TRYPSIN | | 分子名称: | CALCIUM ION, PORCINE E-TRYPSIN | | 著者 | Huang, Q, Wang, Z, Li, Y, Liu, S, Tang, Y. | | 登録日 | 1994-06-07 | | 公開日 | 1995-02-07 | | 最終更新日 | 2024-11-20 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Refined 1.8 A resolution crystal structure of the porcine epsilon-trypsin.
Biochim.Biophys.Acta, 1209, 1994
|
|
3EP3
 
 | | Human AdoMetDC D174N mutant with no putrescine bound | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, PYRUVIC ACID, S-adenosylmethionine decarboxylase alpha chain, ... | | 著者 | Bale, S, Lopez, M.M, Makhatadze, G.I, Fang, Q, Pegg, A.E, Ealick, S.E. | | 登録日 | 2008-09-29 | | 公開日 | 2008-12-23 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.84 Å) | | 主引用文献 | Structural Basis for Putrescine Activation of Human S-Adenosylmethionine Decarboxylase.
Biochemistry, 47, 2008
|
|
6UIL
 
 | | Crystal Structure of Danio rerio Histone Deacetylase 10 in Complex with 7-[(3-aminopropyl)amino]-1,1,1-trifluoroheptan-2-one | | 分子名称: | 7-[(3-aminopropyl)amino]-1,1,1-trifluoroheptane-2,2-diol, PHOSPHATE ION, POTASSIUM ION, ... | | 著者 | Herbst-Gervasoni, C.J, Christianson, D.W. | | 登録日 | 2019-10-01 | | 公開日 | 2019-12-04 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.85 Å) | | 主引用文献 | Binding ofN8-Acetylspermidine Analogues to Histone Deacetylase 10 Reveals Molecular Strategies for Blocking Polyamine Deacetylation.
Biochemistry, 58, 2019
|
|
3ESJ
 
 | | Crystal structure of 2C-methyl-D-erythritol 2,4-clycodiphosphate synthase complexed with ligand | | 分子名称: | 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase, 4-amino-1-[(2S,4aR,6R,7R,7aS)-2,7-dihydroxy-2-oxidotetrahydro-4H-furo[3,2-d][1,3,2]dioxaphosphinin-6-yl]pyrimidin-2(1H)-one, GERANYL DIPHOSPHATE, ... | | 著者 | Hunter, W.N, Ramsden, N.L. | | 登録日 | 2008-10-06 | | 公開日 | 2009-08-25 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | A structure-based approach to ligand discovery for 2C-methyl-D-erythritol-2,4-cyclodiphosphate synthase: a target for antimicrobial therapy
J.Med.Chem., 52, 2009
|
|
4NC6
 
 | |
