4FMQ
 
 | |
3TEI
 
 | |
3PGX
 
 | |
3PXX
 
 | |
1BR3
 
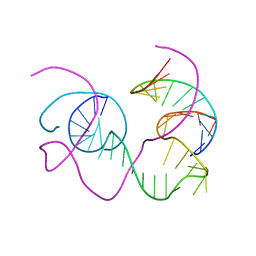 | | CRYSTAL STRUCTURE OF AN 82-NUCLEOTIDE RNA-DNA COMPLEX FORMED BY THE 10-23 DNA ENZYME | | 分子名称: | DNA (10-23 DNA ENZYME), RNA (5'-R(*GP*GP*AP*CP*AP*GP*AP*UP*GP*GP*GP*AP*G)-3') | | 著者 | Nowakowski, J, Shim, P.J, Prasad, G.S, Stout, C.D, Joyce, G.F. | | 登録日 | 1998-08-13 | | 公開日 | 1999-02-23 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Crystal structure of an 82-nucleotide RNA-DNA complex formed by the 10-23 DNA enzyme.
Nat.Struct.Biol., 6, 1999
|
|
2MC3
 
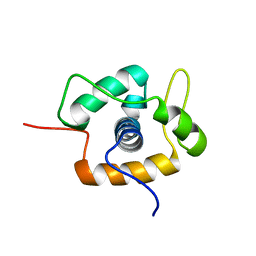 | |
7KTU
 
 | |
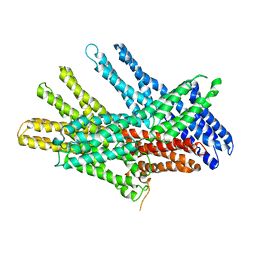
7KTV
 
 | |
7KTT
 
 | |
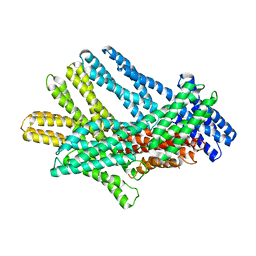
7KTW
 
 | |
6UZK
 
 | |
1KH6
 
 | | Crystal Structure of an RNA Tertiary Domain Essential to HCV IRES-mediated Translation Initiation. | | 分子名称: | JIIIabc | | 著者 | Kieft, J.S, Zhou, K, Grech, A, Jubin, R, Doudna, J.A. | | 登録日 | 2001-11-29 | | 公開日 | 2002-04-26 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Crystal structure of an RNA tertiary domain essential to HCV IRES-mediated translation initiation.
Nat.Struct.Biol., 9, 2002
|
|
1ELG
 
 | | NATURE OF THE INACTIVATION OF ELASTASE BY N-PEPTIDYL-O-AROYL HYDROXYLAMINE AS A FUNCTION OF PH | | 分子名称: | (TERT-BUTYLOXYCARBONYL)-ALANYL-ALANYL-AMINE, CALCIUM ION, PORCINE PANCREATIC ELASTASE | | 著者 | Ding, X, Rasmussen, B, Demuth, H.-U, Ringe, D, Steinmetz, A.C.U. | | 登録日 | 1995-03-13 | | 公開日 | 1995-07-10 | | 最終更新日 | 2024-06-05 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Nature of the inactivation of elastase by N-peptidyl-O-aroyl hydroxylamine as a function of pH.
Biochemistry, 34, 1995
|
|
4GJ1
 
 | | Crystal structure of 1-(5-phosphoribosyl)-5-[(5-phosphoribosylamino)methylideneamino] imidazole-4-carboxamide isomerase (hisA). | | 分子名称: | 1-(5-phosphoribosyl)-5-[(5-phosphoribosylamino)methylideneamino] imidazole-4-carboxamide isomerase | | 著者 | Nocek, B, Gu, M, Kwon, K, Anderson, W.F, Joachimiak, A, CSGID, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2012-08-09 | | 公開日 | 2012-08-22 | | 最終更新日 | 2018-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.152 Å) | | 主引用文献 | Crystal structure of 1-(5-phosphoribosyl)-5-[(5-phosphoribosylamino)methylideneamino] imidazole-4-carboxamide isomerase (hisA).
To be Published
|
|
3N1B
 
 | | C-terminal domain of Vps54 subunit of the GARP complex | | 分子名称: | Vacuolar protein sorting-associated protein 54 | | 著者 | Perez-Victoria, F.J, Abascal-Palacios, G, Tascon, I, Kajava, A, Pioro, E.P, Bonifacino, J.S, Hierro, A. | | 登録日 | 2010-05-15 | | 公開日 | 2010-07-14 | | 最終更新日 | 2017-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.398 Å) | | 主引用文献 | Structural basis for the wobbler mouse neurodegenerative disorder caused by mutation in the Vps54 subunit of the GARP complex.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
5W32
 
 | |
5W2Z
 
 | |
5W31
 
 | |
8I4O
 
 | | Design of a split green fluorescent protein for sensing and tracking an beta-amyloid | | 分子名称: | Beta-amyloid, Split Green flourescent protein | | 著者 | Taegeun, Y, Jinsu, L, Jungmin, Y, Jungmin, C, Wondo, H, Song, J.J, Haksung, K. | | 登録日 | 2023-01-20 | | 公開日 | 2023-11-29 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Engineering of a Fluorescent Protein for a Sensing of an Intrinsically Disordered Protein through Transition in the Chromophore State.
Jacs Au, 3, 2023
|
|
2H3L
 
 | | Crystal Structure of ERBIN PDZ | | 分子名称: | LAP2 protein | | 著者 | Appleton, B.A, Zhang, Y, Wu, P, Yin, J.P, Hunziker, W, Skelton, N.J, Sidhu, S.S, Wiesmann, C. | | 登録日 | 2006-05-22 | | 公開日 | 2006-06-13 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1 Å) | | 主引用文献 | Comparative structural analysis of the Erbin PDZ domain and the first PDZ domain of ZO-1. Insights into determinants of PDZ domain specificity.
J.Biol.Chem., 281, 2006
|
|
2TMA
 
 | |
6OV2
 
 | |
2B6O
 
 | | Electron crystallographic structure of lens Aquaporin-0 (AQP0) (lens MIP) at 1.9A resolution, in a closed pore state | | 分子名称: | 1,2-DIMYRISTOYL-RAC-GLYCERO-3-PHOSPHOCHOLINE, Lens fiber major intrinsic protein | | 著者 | Gonen, T, Cheng, Y, Sliz, P, Hiroaki, Y, Fujiyoshi, Y, Harrison, S.C, Walz, T. | | 登録日 | 2005-10-03 | | 公開日 | 2005-12-06 | | 最終更新日 | 2023-08-23 | | 実験手法 | ELECTRON CRYSTALLOGRAPHY (1.9 Å) | | 主引用文献 | Lipid-protein interactions in double-layered two-dimensional AQP0 crystals.
Nature, 438, 2005
|
|
6OV3
 
 | |
2B6P
 
 | | X-ray structure of lens Aquaporin-0 (AQP0) (lens MIP) in an open pore state | | 分子名称: | Lens fiber major intrinsic protein | | 著者 | Gonen, T, Cheng, Y, Sliz, P, Hiroaki, Y, Fujiyoshi, Y, Harrison, S.C, Walz, T. | | 登録日 | 2005-10-03 | | 公開日 | 2005-12-06 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Lipid-protein interactions in double-layered two-dimensional AQP0 crystals.
Nature, 438, 2005
|
|
