4BDC
 
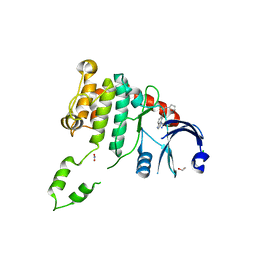 | | Fragment-based screening identifies a new area for inhibitor binding to checkpoint kinase 2 (CHK2) | | 分子名称: | 1,2-ETHANEDIOL, CHLORIDE ION, N-(furan-2-ylmethyl)quinoxaline-6-carboxamide, ... | | 著者 | Silva-Santisteban, M.C, Westwood, I.M, Boxall, K, Brown, N, Peacock, S, McAndrew, C, Barrie, E, Richards, M, Mirza, A, Oliver, A.W, Burke, R, Hoelder, S, Jones, K, Aherne, G.W, Blagg, J, Collins, I, Garrett, M.D, van Montfort, R.L.M. | | 登録日 | 2012-10-05 | | 公開日 | 2013-06-26 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Fragment-Based Screening Maps Inhibitor Interactions in the ATP-Binding Site of Checkpoint Kinase 2.
Plos One, 8, 2013
|
|
3D0Q
 
 | | Crystal structure of calG3 from Micromonospora echinospora determined in space group I222 | | 分子名称: | 3[N-MORPHOLINO]PROPANE SULFONIC ACID, Protein CalG3 | | 著者 | Bitto, E, Singh, S, Bingman, C.A, Wesenberg, G.E, Phillips Jr, G.N. | | 登録日 | 2008-05-02 | | 公開日 | 2008-06-24 | | 最終更新日 | 2017-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.79 Å) | | 主引用文献 | Biochemical and structural insights of the early glycosylation steps in calicheamicin biosynthesis.
Chem.Biol., 15, 2008
|
|
5KM6
 
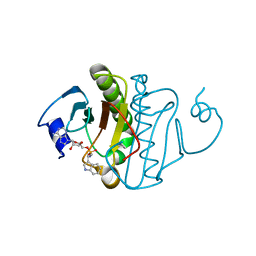 | | Human Histidine Triad Nucleotide Binding Protein 1 (hHint1) H112N mutant Ara-A nucleoside phosphoramidate substrate complex | | 分子名称: | Histidine triad nucleotide-binding protein 1, [(2~{R},3~{S},4~{S},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-~{N}-[2-(1~{H}-indol-3-yl)ethyl]phosphonamidic acid | | 著者 | Maize, K.M, Finzel, B.C. | | 登録日 | 2016-06-26 | | 公開日 | 2017-06-28 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | A Crystal Structure Based Guide to the Design of Human Histidine Triad Nucleotide Binding Protein 1 (hHint1) Activated ProTides.
Mol. Pharm., 14, 2017
|
|
4AXE
 
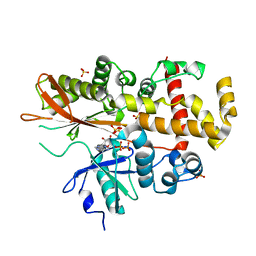 | | Inositol 1,3,4,5,6-pentakisphosphate 2-kinase in complex with ADP | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, INOSITOL-PENTAKISPHOSPHATE 2-KINASE, SULFATE ION, ... | | 著者 | I Banos-Sanz, J, Sanz-Aparicio, J, Gonzalez, B. | | 登録日 | 2012-06-12 | | 公開日 | 2012-07-04 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Conformational Changes Undergone by Inositol 1,3,4,5,6-Pentakisphosphate 2-Kinase Upon Substrate Binding: The Role of N-Lobe and Enantiomeric Substrate Preference
J.Biol.Chem., 287, 2012
|
|
5KM2
 
 | |
4AXD
 
 | | Inositol 1,3,4,5,6-pentakisphosphate 2-kinase in complex with AMPPNP | | 分子名称: | CITRIC ACID, INOSITOL-PENTAKISPHOSPHATE 2-KINASE, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | 著者 | I Banos-Sanz, J, Sanz-Aparicio, J, Gonzalez, B. | | 登録日 | 2012-06-12 | | 公開日 | 2012-07-04 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Conformational Changes Undergone by Inositol 1,3,4,5,6-Pentakisphosphate 2-Kinase Upon Substrate Binding: The Role of N-Lobe and Enantiomeric Substrate Preference
J.Biol.Chem., 287, 2012
|
|
4AXC
 
 | | Inositol 1,3,4,5,6-pentakisphosphate 2-kinase apo form | | 分子名称: | GLYCEROL, INOSITOL-PENTAKISPHOSPHATE 2-KINASE, SULFATE ION, ... | | 著者 | I Banos-Sanz, J, Sanz-Aparicio, J, Gonzalez, B. | | 登録日 | 2012-06-12 | | 公開日 | 2012-07-04 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Conformational Changes Undergone by Inositol 1,3,4,5,6-Pentakisphosphate 2-Kinase Upon Substrate Binding: The Role of N-Lobe and Enantiomeric Substrate Preference
J.Biol.Chem., 287, 2012
|
|
6A51
 
 | | Novel Regulators CheP and CheQ Specifically Control Chemotaxis Core Gene cheVAW Transcription in Bacterial Pathogen Campylobacter jejuni | | 分子名称: | CheQ | | 著者 | Lu, G, Gao, B, Cha, G, Chen, Z, Mo, R. | | 登録日 | 2018-06-21 | | 公開日 | 2019-06-26 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | The novel regulators CheP and CheQ control the core chemotaxis operon cheVAW in Campylobacter jejuni.
Mol.Microbiol., 111, 2019
|
|
2CZJ
 
 | | Crystal structure of the tRNA domain of tmRNA from Thermus thermophilus HB8 | | 分子名称: | SsrA-binding protein, tmRNA (63-MER) | | 著者 | Bessho, Y, Shibata, R, Sekine, S, Murayama, K, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2005-07-13 | | 公開日 | 2006-10-31 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (3.01 Å) | | 主引用文献 | Structural basis for functional mimicry of long-variable-arm tRNA by transfer-messenger RNA.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
6DMA
 
 | | DHD15_closed | | 分子名称: | DHD15_closed_A, DHD15_closed_B | | 著者 | Bick, M.J, Chen, Z, Baker, D. | | 登録日 | 2018-06-04 | | 公開日 | 2018-12-19 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (3.363 Å) | | 主引用文献 | Programmable design of orthogonal protein heterodimers.
Nature, 565, 2019
|
|
4BDD
 
 | | Fragment-based screening identifies a new area for inhibitor binding to checkpoint kinase 2 (CHK2) | | 分子名称: | 1,2-ETHANEDIOL, 1-(TERT-BUTYL)-3-(QUINOXALIN-6-YL)UREA, NITRATE ION, ... | | 著者 | Silva-Santisteban, M.C, Westwood, I.M, Boxall, K, Brown, N, Peacock, S, McAndrew, C, Barrie, E, Richards, M, Mirza, A, Oliver, A.W, Burke, R, Hoelder, S, Jones, K, Aherne, G.W, Blagg, J, Collins, I, Garrett, M.D, van Montfort, R.L.M. | | 登録日 | 2012-10-05 | | 公開日 | 2013-06-26 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.67 Å) | | 主引用文献 | Fragment-Based Screening Maps Inhibitor Interactions in the ATP-Binding Site of Checkpoint Kinase 2.
Plos One, 8, 2013
|
|
3COP
 
 | |
2CV2
 
 | | Glutamyl-tRNA synthetase from Thermus thermophilus in complex with tRNA(Glu) and an enzyme inhibitor, Glu-AMS | | 分子名称: | CHLORIDE ION, MAGNESIUM ION, O5'-(L-GLUTAMYL-SULFAMOYL)-ADENOSINE, ... | | 著者 | Sekine, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2005-05-31 | | 公開日 | 2006-09-05 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.69 Å) | | 主引用文献 | Structural bases of transfer RNA-dependent amino acid recognition and activation by glutamyl-tRNA synthetase
Structure, 14, 2006
|
|
4BLA
 
 | | Crystal structure of full-length human Suppressor of fused (SUFU) mutant lacking a regulatory subdomain (crystal form II) | | 分子名称: | MALTOSE-BINDING PERIPLASMIC PROTEIN, SUPPRESSOR OF FUSED HOMOLOG | | 著者 | Cherry, A.L, Finta, C, Karlstrom, M, Toftgard, R, Jovine, L. | | 登録日 | 2013-05-02 | | 公開日 | 2013-11-27 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (3.5 Å) | | 主引用文献 | Structural Basis of Sufu-GLI Interaction in Hedgehog Signalling Regulation
Acta Crystallogr.,Sect.D, 69, 2013
|
|
8SZV
 
 | | Crystal structure of pregnane X receptor ligand binding domain complexed with T0901317 analog SJPYT-318 | | 分子名称: | N-[4-(1,1,1,3,3,3-hexafluoro-2-hydroxypropan-2-yl)phenyl]-N-[(4-phenoxyphenyl)methyl]benzenesulfonamide, Nuclear receptor subfamily 1 group I member 2 | | 著者 | Huber, A.D, Poudel, S, Miller, D.J, Li, Y, Chen, T. | | 登録日 | 2023-05-30 | | 公開日 | 2023-10-11 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Ligand flexibility and binding pocket malleability cooperate to allow selective PXR activation by analogs of a promiscuous nuclear receptor ligand.
Structure, 31, 2023
|
|
4BDE
 
 | | Fragment-based screening identifies a new area for inhibitor binding to checkpoint kinase 2 (CHK2) | | 分子名称: | 1,2-ETHANEDIOL, 6-METHYLQUINAZOLIN-4-AMINE, NITRATE ION, ... | | 著者 | Silva-Santisteban, M.C, Westwood, I.M, Boxall, K, Brown, N, Peacock, S, McAndrew, C, Barrie, E, Richards, M, Mirza, A, Oliver, A.W, Burke, R, Hoelder, S, Jones, K, Aherne, G.W, Blagg, J, Collins, I, Garrett, M.D, van Montfort, R.L.M. | | 登録日 | 2012-10-05 | | 公開日 | 2013-06-26 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.55 Å) | | 主引用文献 | Fragment-Based Screening Maps Inhibitor Interactions in the ATP-Binding Site of Checkpoint Kinase 2.
Plos One, 8, 2013
|
|
3CQN
 
 | |
3CS0
 
 | | Crystal structure of DegP24 | | 分子名称: | Periplasmic serine endoprotease DegP, pentapeptide | | 著者 | Krojer, T, Sawa, J, Schaefer, E, Saibil, H.R, Ehrmann, M, Clausen, T. | | 登録日 | 2008-04-08 | | 公開日 | 2008-05-27 | | 最終更新日 | 2019-11-13 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Structural basis for the regulated protease and chaperone function of DegP
Nature, 453, 2008
|
|
3CSB
 
 | |
3CX5
 
 | |
2D5R
 
 | | Crystal Structure of a Tob-hCaf1 Complex | | 分子名称: | CCR4-NOT transcription complex subunit 7, Tob1 protein | | 著者 | Horiuchi, M, Suzuki, N.N, Muroya, N, Takahasi, K, Nishida, M, Yoshida, Y, Ikematsu, N, Nakamura, T, Kawamura-Tsuzuku, J, Yamamoto, T, Inagaki, F. | | 登録日 | 2005-11-04 | | 公開日 | 2006-12-12 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structural basis for the antiproliferative activity of the Tob-hCaf1 complex.
J.Biol.Chem., 284, 2009
|
|
3D1B
 
 | |
3D36
 
 | | How to Switch Off a Histidine Kinase: Crystal Structure of Geobacillus stearothermophilus KinB with the Inhibitor Sda | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | 著者 | Bick, M.J, Lamour, V, Rajashankar, K.R, Gordiyenko, Y, Robinson, C.V, Darst, S.A. | | 登録日 | 2008-05-09 | | 公開日 | 2009-01-13 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.03 Å) | | 主引用文献 | How to switch off a histidine kinase: crystal structure of Geobacillus stearothermophilus KinB with the inhibitor Sda
J.Mol.Biol., 386, 2009
|
|
4BL7
 
 | |
5KLY
 
 | |
