2BGP
 
 | | Mannan Binding Module from Man5C in bound conformation | | 分子名称: | ENDO-B1,4-MANNANASE 5C | | 著者 | Tunnicliffe, R.B, Bolam, D.N, Pell, G, Gilbert, H.J, Williamson, M.P. | | 登録日 | 2005-01-04 | | 公開日 | 2005-03-09 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structure of a Mannan-Specific Family 35 Carbohydrate-Binding Module: Evidence for Significant Conformational Changes Upon Ligand Binding
J.Mol.Biol., 347, 2005
|
|
2BL5
 
 | | Solution structure of the KH-QUA2 region of the Xenopus STAR-GSG Quaking protein. | | 分子名称: | MGC83862 PROTEIN | | 著者 | Maguire, M.L, Guler-Gane, G, Nietlispach, D, Raine, A.R.C, Zorn, A.M, Standart, N, Broadhurst, R.W. | | 登録日 | 2005-03-01 | | 公開日 | 2005-04-14 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution Structure and Backbone Dynamics of the Kh-Qua2 Region of the Xenopus Star/Gsg Quaking Protein
J.Mol.Biol., 348, 2005
|
|
2BLN
 
 | | N-terminal formyltransferase domain of ArnA in complex with N-5- formyltetrahydrofolate and UMP | | 分子名称: | ACETATE ION, N-{[4-({[(6R)-2-amino-5-formyl-4-oxo-1,4,5,6,7,8-hexahydropteridin-6-yl]methyl}amino)phenyl]carbonyl}-L-glutamic acid, PROTEIN YFBG, ... | | 著者 | Williams, G.J, Breazeale, S.D, Raetz, C.R.H, Naismith, J.H. | | 登録日 | 2005-03-07 | | 公開日 | 2005-04-08 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.2 Å) | | 主引用文献 | Structure and Function of Both Domains of Arna, a Dual Function Decarboxylase and a Formyltransferase, Involved in 4-Amino-4-Deoxy-L-Arabinose Biosynthesis.
J.Biol.Chem., 280, 2005
|
|
2BHE
 
 | | HUMAN CYCLIN DEPENDENT PROTEIN KINASE 2 IN COMPLEX WITH THE INHIBITOR 5-BROMO-INDIRUBINE | | 分子名称: | (2Z)-5'-BROMO-2,3'-BIINDOLE-2',3(1H,1'H)-DIONE AMMONIATE, CELL DIVISION PROTEIN KINASE 2 | | 著者 | Schaefer, M, Jautelat, R, Brumby, T, Briem, H, Eisenbrand, G, Schwahn, S, Krueger, M, Luecking, U, Prien, O, Siemeister, G. | | 登録日 | 2005-01-10 | | 公開日 | 2005-03-09 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | From the Insoluble Dye Indirubin Towards Highly Active, Soluble Cdk2-Inhibitors
Chembiochem, 6, 2005
|
|
2BUN
 
 | | Solution structure of the BLUF domain of AppA 5-125 | | 分子名称: | APPA, FLAVIN-ADENINE DINUCLEOTIDE | | 著者 | Grinstead, J.S, Hsu, S.-T, Laan, W, Bonvin, A.M.J.J, Hellingwerf, K.J, Boelens, R, Kaptein, R. | | 登録日 | 2005-06-15 | | 公開日 | 2005-12-07 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The solution structure of the AppA BLUF domain: insight into the mechanism of light-induced signaling.
Chembiochem, 7, 2006
|
|
2BO5
 
 | | Bovine oligomycin sensitivity conferral protein N-terminal domain | | 分子名称: | ATP SYNTHASE OLIGOMYCIN SENSITIVITY CONFERRAL PROTEIN | | 著者 | Carbajo, R.J, Kellas, F.A, Runswick, M.J, Montgomery, M.G, Walker, J.E, Neuhaus, D. | | 登録日 | 2005-04-07 | | 公開日 | 2005-08-17 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structure of the F1-binding domain of the stator of bovine F1Fo-ATPase and how it binds an alpha-subunit.
J. Mol. Biol., 351, 2005
|
|
2C52
 
 | | Structural diversity in CBP p160 complexes | | 分子名称: | CREB-BINDING PROTEIN, NUCLEAR RECEPTOR COACTIVATOR 1 | | 著者 | Waters, L.C, Yue, B, Veverka, V, Renshaw, P.S, Bramham, J, Matsuda, S, Frenkiel, T, Kelly, G, Muskett, F.W, Carr, M.D, Heery, D.M. | | 登録日 | 2005-10-25 | | 公開日 | 2006-03-15 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural diversity in p160/CREB-binding protein coactivator complexes.
J. Biol. Chem., 281, 2006
|
|
2BTG
 
 | |
7QEQ
 
 | | human Connexin 26 dodecamer at 90mmHg PCO2, pH7.4 | | 分子名称: | DODECYL-BETA-D-MALTOSIDE, Gap junction beta-2 protein, PHOSPHATIDYLETHANOLAMINE | | 著者 | Brotherton, D.H, Cameron, A.D, Savva, C.G, Ragan, T.J. | | 登録日 | 2021-12-03 | | 公開日 | 2022-03-30 | | 最終更新日 | 2022-05-18 | | 実験手法 | ELECTRON MICROSCOPY (1.9 Å) | | 主引用文献 | Conformational changes and CO 2 -induced channel gating in connexin26.
Structure, 30, 2022
|
|
7QET
 
 | | human Connexin 26 dodecamer at 20mmHg PCO2, pH7.4 | | 分子名称: | DODECYL-BETA-D-MALTOSIDE, Gap junction beta-2 protein, PHOSPHATIDYLETHANOLAMINE | | 著者 | Brotherton, D.H, Cameron, A.D, Savva, C.G, Ragan, T.J. | | 登録日 | 2021-12-03 | | 公開日 | 2022-03-30 | | 最終更新日 | 2022-05-18 | | 実験手法 | ELECTRON MICROSCOPY (2.1 Å) | | 主引用文献 | Conformational changes and CO 2 -induced channel gating in connexin26.
Structure, 30, 2022
|
|
7QP6
 
 | | Structure of the human 48S initiation complex in open state (h48S AUG open) | | 分子名称: | 18S rRNA, 40S ribosomal protein S10, 40S ribosomal protein S11, ... | | 著者 | Yi, S.-H, Petrychenko, V, Schliep, J.E, Goyal, A, Linden, A, Chari, A, Urlaub, H, Stark, H, Rodnina, M.V, Adio, S, Fischer, N. | | 登録日 | 2022-01-03 | | 公開日 | 2022-05-11 | | 最終更新日 | 2024-04-24 | | 実験手法 | ELECTRON MICROSCOPY (4.7 Å) | | 主引用文献 | Conformational rearrangements upon start codon recognition in human 48S translation initiation complex.
Nucleic Acids Res., 50, 2022
|
|
7QP7
 
 | | Structure of the human 48S initiation complex in closed state (h48S AUG closed) | | 分子名称: | 18S rRNA, 40S ribosomal protein S10, 40S ribosomal protein S11, ... | | 著者 | Yi, S.-H, Petrychenko, V, Schliep, J.E, Goyal, A, Linden, A, Chari, A, Urlaub, H, Stark, H, Rodnina, M.V, Adio, S, Fischer, N. | | 登録日 | 2022-01-03 | | 公開日 | 2022-05-11 | | 最終更新日 | 2024-04-24 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Conformational rearrangements upon start codon recognition in human 48S translation initiation complex.
Nucleic Acids Res., 50, 2022
|
|
7QSL
 
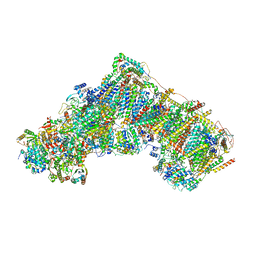 | | Bovine complex I in lipid nanodisc, Active-apo | | 分子名称: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, Acyl carrier protein, ... | | 著者 | Chung, I, Bridges, H.R, Hirst, J. | | 登録日 | 2022-01-13 | | 公開日 | 2022-05-25 | | 最終更新日 | 2022-09-28 | | 実験手法 | ELECTRON MICROSCOPY (2.76 Å) | | 主引用文献 | Cryo-EM structures define ubiquinone-10 binding to mitochondrial complex I and conformational transitions accompanying Q-site occupancy.
Nat Commun, 13, 2022
|
|
7QSM
 
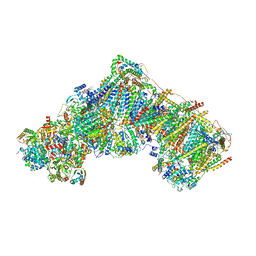 | | Bovine complex I in lipid nanodisc, Deactive-ligand (composite) | | 分子名称: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, Acyl carrier protein, ... | | 著者 | Chung, I, Bridges, H.R, Hirst, J. | | 登録日 | 2022-01-13 | | 公開日 | 2022-05-25 | | 最終更新日 | 2022-09-28 | | 実験手法 | ELECTRON MICROSCOPY (2.3 Å) | | 主引用文献 | Cryo-EM structures define ubiquinone-10 binding to mitochondrial complex I and conformational transitions accompanying Q-site occupancy.
Nat Commun, 13, 2022
|
|
7QSN
 
 | | Bovine complex I in lipid nanodisc, Deactive-apo | | 分子名称: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, Acyl carrier protein, ... | | 著者 | Chung, I, Bridges, H.R, Hirst, J. | | 登録日 | 2022-01-13 | | 公開日 | 2022-05-25 | | 最終更新日 | 2022-09-28 | | 実験手法 | ELECTRON MICROSCOPY (2.81 Å) | | 主引用文献 | Cryo-EM structures define ubiquinone-10 binding to mitochondrial complex I and conformational transitions accompanying Q-site occupancy.
Nat Commun, 13, 2022
|
|
7QSK
 
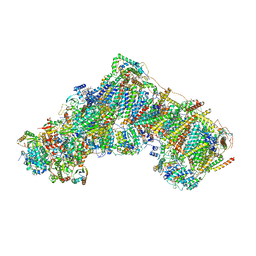 | | Bovine complex I in lipid nanodisc, Active-Q10 | | 分子名称: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, Acyl carrier protein, ... | | 著者 | Chung, I, Bridges, H.R, Hirst, J. | | 登録日 | 2022-01-13 | | 公開日 | 2022-05-25 | | 最終更新日 | 2022-09-28 | | 実験手法 | ELECTRON MICROSCOPY (2.84 Å) | | 主引用文献 | Cryo-EM structures define ubiquinone-10 binding to mitochondrial complex I and conformational transitions accompanying Q-site occupancy.
Nat Commun, 13, 2022
|
|
7QSO
 
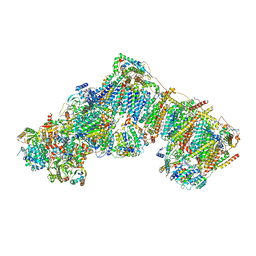 | | Bovine complex I in lipid nanodisc, State 3 (Slack) | | 分子名称: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, Acyl carrier protein, ... | | 著者 | Chung, I, Bridges, H.R, Hirst, J. | | 登録日 | 2022-01-13 | | 公開日 | 2022-05-25 | | 最終更新日 | 2022-09-28 | | 実験手法 | ELECTRON MICROSCOPY (3.02 Å) | | 主引用文献 | Cryo-EM structures define ubiquinone-10 binding to mitochondrial complex I and conformational transitions accompanying Q-site occupancy.
Nat Commun, 13, 2022
|
|
7QEU
 
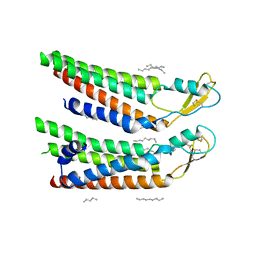 | | human Connexin 26 at 55mmHg PCO2, pH7.4: two masked subunits, class B | | 分子名称: | DODECYL-BETA-D-MALTOSIDE, Gap junction beta-2 protein, PHOSPHATIDYLETHANOLAMINE | | 著者 | Brotherton, D.H, Cameron, A.D, Savva, C.G, Ragan, T.J. | | 登録日 | 2021-12-03 | | 公開日 | 2022-06-15 | | 実験手法 | ELECTRON MICROSCOPY (2.7 Å) | | 主引用文献 | Conformational changes and CO 2 -induced channel gating in connexin26.
Structure, 30, 2022
|
|
7QEO
 
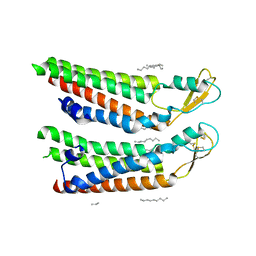 | | human Connexin 26 at 55mm Hg PCO2, pH7.4: two masked subunits, class C | | 分子名称: | DODECYL-BETA-D-MALTOSIDE, Gap junction beta-2 protein, PHOSPHATIDYLETHANOLAMINE | | 著者 | Brotherton, D.H, Cameron, A.D, Savva, C.G, Ragan, T.J. | | 登録日 | 2021-12-03 | | 公開日 | 2022-06-22 | | 最終更新日 | 2024-10-16 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Conformational changes and CO 2 -induced channel gating in connexin26.
Structure, 30, 2022
|
|
7CM5
 
 | |
7QQ8
 
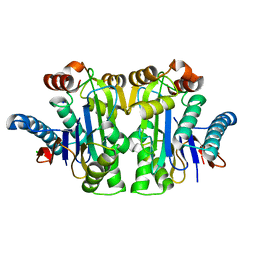 | | Structure of E.coli Class 2 L-asparaginase EcAIII, mutant RDM1-8 (G206Y, R207Q, D210P, S211T) | | 分子名称: | Beta-aspartyl-peptidase, CHLORIDE ION, SODIUM ION | | 著者 | Loch, J.I, Kadziolka, K, Jaskolski, M. | | 登録日 | 2022-01-06 | | 公開日 | 2022-07-13 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structural and biophysical studies of new L-asparaginase variants: lessons from random mutagenesis of the prototypic Escherichia coli Ntn-amidohydrolase.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7CHK
 
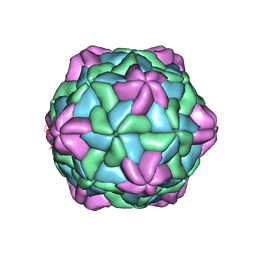 | | Cryo-EM Structure of Apple Latent Spherical Virus (ALSV) | | 分子名称: | VP20 protein, VP24 protein, VP25 protein | | 著者 | Naitow, H, Hamaguchi, T, Maki-Yonekura, S, Isogai, M, Yoshikawa, N, Yonekura, K. | | 登録日 | 2020-07-06 | | 公開日 | 2020-11-04 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (2.87 Å) | | 主引用文献 | Apple latent spherical virus structure with stable capsid frame supports quasi-stable protrusions expediting genome release.
Commun Biol, 3, 2020
|
|
7Q67
 
 | | Cryo-em structure of the Nup98 fibril polymorph 4 | | 分子名称: | Nuclear pore complex protein Nup98 | | 著者 | Ibanez de Opakua, A, Geraets, J.A, Frieg, B, Dienemann, C, Savastano, A, Rankovic, M, Cima-Omori, M.-S, Schroeder, G.F, Zweckstetter, M. | | 登録日 | 2021-11-05 | | 公開日 | 2022-10-12 | | 最終更新日 | 2024-07-17 | | 実験手法 | ELECTRON MICROSCOPY (3.37 Å) | | 主引用文献 | Molecular interactions of FG nucleoporin repeats at high resolution.
Nat.Chem., 14, 2022
|
|
7Q65
 
 | | Cryo-em structure of the Nup98 fibril polymorph 2 | | 分子名称: | Nuclear pore complex protein Nup98 | | 著者 | Ibanez de Opakua, A, Geraets, J.A, Frieg, B, Dienemann, C, Savastano, A, Rankovic, M, Cima-Omori, M.-S, Schroeder, G.F, Zweckstetter, M. | | 登録日 | 2021-11-05 | | 公開日 | 2022-10-12 | | 最終更新日 | 2024-07-17 | | 実験手法 | ELECTRON MICROSCOPY (3.32 Å) | | 主引用文献 | Molecular interactions of FG nucleoporin repeats at high resolution.
Nat.Chem., 14, 2022
|
|
7QVI
 
 | | Fiber-forming RubisCO derived from ancestral sequence reconstruction and rational engineering | | 分子名称: | 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE, MAGNESIUM ION, RubisCO large subunit | | 著者 | Schulz, L, Zarzycki, J, Prinz, S, Schuller, J.M, Erb, T.J, Hochberg, G.K.A. | | 登録日 | 2022-01-21 | | 公開日 | 2022-10-12 | | 最終更新日 | 2022-10-26 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Evolution of increased complexity and specificity at the dawn of form I Rubiscos.
Science, 378, 2022
|
|
