8D20
 
 | |
8D1S
 
 | | Crystal structure of Plasmodium falciparum GRP78 in complex with Toyocamycin | | 分子名称: | 4-amino-7-(beta-D-ribofuranosyl)-7H-pyrrolo[2,3-d]pyrimidine-5-carbonitrile, Chaperone DnaK, SULFATE ION | | 著者 | Mrozek, A, Chen, Y, Antoshchenko, T, Park, H.W. | | 登録日 | 2022-05-27 | | 公開日 | 2023-05-31 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | A non-traditional crystal-based compound screening method targeting the ATP binding site of Plasmodium falciparum GRP78 for identification of novel nucleoside analogues.
Front Mol Biosci, 9, 2022
|
|
4AM6
 
 | | C-TERMINAL DOMAIN OF ACTIN-RELATED PROTEIN ARP8 FROM S. CEREVISIAE | | 分子名称: | ACTIN-LIKE PROTEIN ARP8, SULFATE ION | | 著者 | Wuerges, J, Saravanan, M, Bose, D, Cook, N.J, Zhang, X, Wigley, D.B. | | 登録日 | 2012-03-07 | | 公開日 | 2012-12-12 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Interactions between the Nucleosome Histone Core and Arp8 in the Ino80 Chromatin Remodeling Complex.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4AM7
 
 | | ADP-BOUND C-TERMINAL DOMAIN OF ACTIN-RELATED PROTEIN ARP8 FROM S. CEREVISIAE | | 分子名称: | ACTIN-LIKE PROTEIN ARP8, ADENOSINE-5'-DIPHOSPHATE | | 著者 | Wuerges, J, Saravanan, M, Bose, D, Cook, N.J, Zhang, X, Wigley, D.B. | | 登録日 | 2012-03-07 | | 公開日 | 2012-12-12 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (3.25 Å) | | 主引用文献 | Interactions between the Nucleosome Histone Core and Arp8 in the Ino80 Chromatin Remodeling Complex.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
8D22
 
 | |
9CB7
 
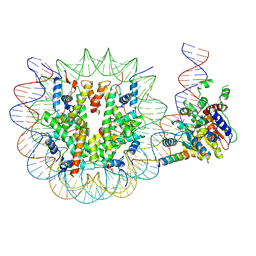 | | DeltaNhp10 INO80 bound to S.c 0/40 nucleosome, Ino80-Nucleosome | | 分子名称: | Chromatin-remodeling ATPase INO80, DNA (227-MER), Histone H2A.1, ... | | 著者 | Wu, H, Kaur, U, Narlikar, G.J, Cheng, Y.F. | | 登録日 | 2024-06-18 | | 公開日 | 2025-07-16 | | 実験手法 | ELECTRON MICROSCOPY (4.04 Å) | | 主引用文献 | Autoinhibition imposed by a large conformational switch of INO80 regulates nucleosome positioning
Science, 2025
|
|
4A0X
 
 | | Structure of the global transcription regulator FapR from Staphylococcus aureus | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ACETATE ION, TRANSCRIPTION FACTOR FAPR, ... | | 著者 | Albanesi, D, Guerin, M.E, Buschiazzo, A, de Mendoza, D, Alzari, P.M. | | 登録日 | 2011-09-13 | | 公開日 | 2012-09-26 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Structural Basis for Feed-Forward Transcriptional Regulation of Membrane Lipid Homeostasis in Staphylococcus Aureus.
Plos Pathog., 9, 2013
|
|
8D5D
 
 | |
8D88
 
 | |
6TJ6
 
 | |
6TJ5
 
 | |
7E5T
 
 | | Crystal structure of Fsa2 | | 分子名称: | Diels-Alderase fsa2, ETHANOL, PENTAETHYLENE GLYCOL, ... | | 著者 | Fujiyama, K, Kato, N, Kinugasa, K, Hino, T, Takahashi, S, Nagano, S. | | 登録日 | 2021-02-20 | | 公開日 | 2021-06-30 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.16977525 Å) | | 主引用文献 | Molecular Basis for Two Stereoselective Diels-Alderases that Produce Decalin Skeletons*.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
3F4M
 
 | | Crystal structure of TIPE2 | | 分子名称: | CHLORIDE ION, Tumor necrosis factor, alpha-induced protein 8-like protein 2 | | 著者 | Zhang, X, Wang, J, Shi, Y. | | 登録日 | 2008-11-01 | | 公開日 | 2009-03-17 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.696 Å) | | 主引用文献 | Crystal structure of TIPE2 provides insights into immune homeostasis
Nat.Struct.Mol.Biol., 16, 2009
|
|
6Y1M
 
 | | Crystal structure of the paraoxon-modified A.17 antibody FAB fragment - L47K mutant | | 分子名称: | DIETHYL PHOSPHONATE, FAB A.17 L47K mutant HEAVY CHAIN, FAB A.17 L47K mutant Light CHAIN | | 著者 | Chatziefthimiou, S, Mokrushina, Y, Smirnov, I, Gabibov, A, Wilmanns, M. | | 登録日 | 2020-02-12 | | 公開日 | 2020-09-16 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Multiscale computation delivers organophosphorus reactivity and stereoselectivity to immunoglobulin scavengers.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6ZYI
 
 | |
6ZYJ
 
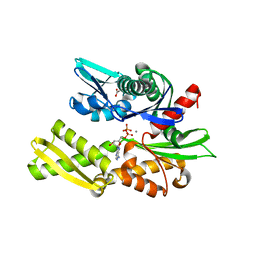 | | Crystal structure of Hsc70 ATPase domain in complex with ADP and calcium | | 分子名称: | ACETATE ION, ADENOSINE-5'-DIPHOSPHATE, CALCIUM ION, ... | | 著者 | Yan, Y, Preissler, S, Ron, D. | | 登録日 | 2020-08-01 | | 公開日 | 2020-12-16 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Calcium depletion challenges endoplasmic reticulum proteostasis by destabilising BiP-substrate complexes.
Elife, 9, 2020
|
|
4A0Z
 
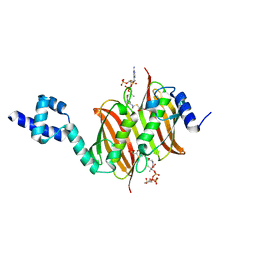 | | Structure of the global transcription regulator FapR from Staphylococcus aureus in complex with malonyl-CoA | | 分子名称: | MALONYL-COENZYME A, TRANSCRIPTION FACTOR FAPR | | 著者 | Albanesi, D, Guerin, M.E, Buschiazzo, A, de Mendoza, D, Alzari, P.M. | | 登録日 | 2011-09-13 | | 公開日 | 2012-09-26 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structural Basis for Feed-Forward Transcriptional Regulation of Membrane Lipid Homeostasis in Staphylococcus Aureus.
Plos Pathog., 9, 2013
|
|
7A4V
 
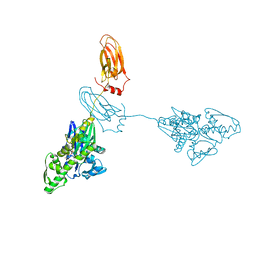 | |
6ZYH
 
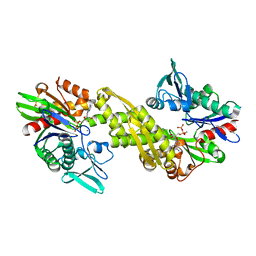 | |
3VOK
 
 | | X-ray Crystal Structure of Wild Type HrtR in the Apo Form with the Target DNA. | | 分子名称: | 5'-D(*AP*TP*GP*AP*CP*AP*CP*TP*GP*TP*GP*TP*CP*AP*T)-3', Transcriptional regulator | | 著者 | Sawai, H, Sugimoto, H, Shiro, Y, Aono, S. | | 登録日 | 2012-01-27 | | 公開日 | 2012-07-25 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural Basis for the Transcriptional Regulation of Heme Homeostasis in Lactococcus lactis.
J.Biol.Chem., 287, 2012
|
|
3VOX
 
 | |
7L1G
 
 | | PRMT5-MEP50 Complexed with SAM | | 分子名称: | 1,2-ETHANEDIOL, CHLORIDE ION, Methylosome protein 50, ... | | 著者 | Palte, R.L. | | 登録日 | 2020-12-14 | | 公開日 | 2021-04-14 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (2.47 Å) | | 主引用文献 | Development of a Flexible and Robust Synthesis of Tetrahydrofuro[3,4- b ]furan Nucleoside Analogues.
J.Org.Chem., 86, 2021
|
|
8UQE
 
 | | Crystal structure of RNF168 (RING)-UbcH5c fused to H2A-H2B via a 26-residue linker (RING not modeled in density) | | 分子名称: | E3 ubiquitin-protein ligase RNF168,Ubiquitin-conjugating enzyme E2 D3,Histone H2B type 2-E,Histone H2A type 1-B/E | | 著者 | Hu, Q, Botuyan, M.V, Mer, G. | | 登録日 | 2023-10-23 | | 公開日 | 2024-01-17 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (3.562 Å) | | 主引用文献 | Mechanisms of RNF168 nucleosome recognition and ubiquitylation.
Mol.Cell, 84, 2024
|
|
8UQA
 
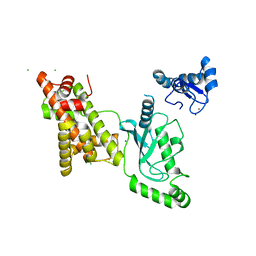 | | Crystal structure of RNF168 (RING)-UbcH5c fused to H2A-H2B via a 12-residue linker | | 分子名称: | CHLORIDE ION, E3 ubiquitin-protein ligase RNF168,Ubiquitin-conjugating enzyme E2 D3,Histone H2B type 2-E,Histone H2A type 1-B/E, SODIUM ION, ... | | 著者 | Hu, Q, Botuyan, M.V, Mer, G. | | 登録日 | 2023-10-23 | | 公開日 | 2024-01-17 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.049 Å) | | 主引用文献 | Mechanisms of RNF168 nucleosome recognition and ubiquitylation.
Mol.Cell, 84, 2024
|
|
8UQB
 
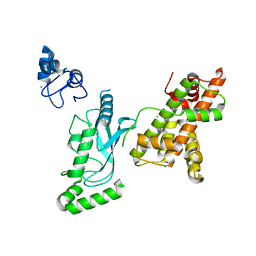 | | Crystal structure of RNF168 (RING)-UbcH5c fused to H2A-H2B via a 20-residue linker (crystallization condition 1) | | 分子名称: | CHLORIDE ION, E3 ubiquitin-protein ligase RNF168,Ubiquitin-conjugating enzyme E2 D3,Histone H2B type 2-E,Histone H2A type 1-B/E, ZINC ION | | 著者 | Hu, Q, Botuyan, M.V, Mer, G. | | 登録日 | 2023-10-23 | | 公開日 | 2024-01-17 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.484 Å) | | 主引用文献 | Mechanisms of RNF168 nucleosome recognition and ubiquitylation.
Mol.Cell, 84, 2024
|
|
