1YKF
 
 | | NADP-DEPENDENT ALCOHOL DEHYDROGENASE FROM THERMOANAEROBIUM BROCKII | | 分子名称: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, NADP-DEPENDENT ALCOHOL DEHYDROGENASE, ZINC ION | | 著者 | Korkhin, Y, Frolow, F. | | 登録日 | 1996-03-25 | | 公開日 | 1998-01-14 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | NADP-dependent bacterial alcohol dehydrogenases: crystal structure, cofactor-binding and cofactor specificity of the ADHs of Clostridium beijerinckii and Thermoanaerobacter brockii.
J.Mol.Biol., 278, 1998
|
|
1YQ5
 
 | | PRD1 vertex protein P5 | | 分子名称: | Minor capsid protein | | 著者 | Merckel, M.C, Huiskonen, J.T, Goldman, A, Bamford, D.H, Tuma, R. | | 登録日 | 2005-02-01 | | 公開日 | 2005-04-26 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | The structure of the bacteriophage PRD1 spike sheds light on the evolution of viral capsid architecture.
Mol.Cell, 18, 2005
|
|
1V1B
 
 | |
1G3G
 
 | | NMR STRUCTURE OF THE FHA1 DOMAIN OF YEAST RAD53 | | 分子名称: | PROTEIN KINASE SPK1 | | 著者 | Yuan, C, Liao, H, Su, M, Yongkiettrakul, S, Byeon, I.-J.L, Tsai, M.-D. | | 登録日 | 2000-10-24 | | 公開日 | 2001-01-10 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structure of the FHA1 domain of yeast Rad53 and identification of binding sites for both FHA1 and its target protein Rad9
J.Mol.Biol., 304, 2000
|
|
1LBE
 
 | | APLYSIA ADP RIBOSYL CYCLASE | | 分子名称: | ADP RIBOSYL CYCLASE | | 著者 | Prasad, G.S, Mcree, D.E, Stura, E.A, Levitt, D.G, Lee, H.C, Stout, C.D. | | 登録日 | 1996-09-18 | | 公開日 | 1997-09-17 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Crystal structure of Aplysia ADP ribosyl cyclase, a homologue of the bifunctional ectozyme CD38.
Nat.Struct.Biol., 3, 1996
|
|
1BDV
 
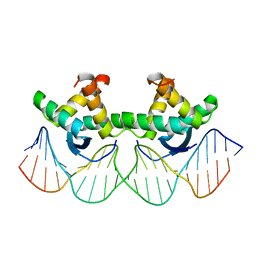 | | ARC FV10 COCRYSTAL | | 分子名称: | DNA (5'-D(*AP*AP*TP*GP*AP*TP*AP*GP*AP*AP*GP*CP*AP*CP*TP*CP*TP*AP*CP*TP*AP*T)-3'), DNA (5'-D(*TP*AP*TP*AP*GP*TP*AP*GP*AP*GP*TP*GP*CP*TP*TP*CP*TP*AP*TP*CP*AP*T)-3'), PROTEIN (ARC FV10 REPRESSOR) | | 著者 | Schildbach, J.F, Karzai, A.W, Raumann, B.E, Sauer, R.T. | | 登録日 | 1998-05-11 | | 公開日 | 1999-01-06 | | 最終更新日 | 2023-08-02 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Origins of DNA-binding specificity: role of protein contacts with the DNA backbone.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
1APN
 
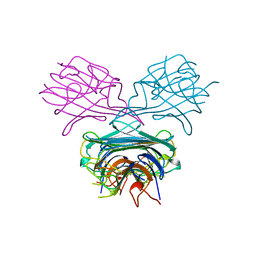 | |
1YLR
 
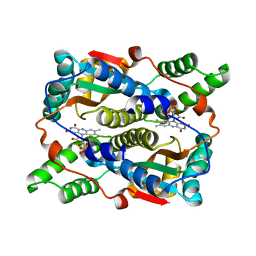 | | The structure of E.coli nitroreductase with bound acetate, crystal form 1 | | 分子名称: | ACETATE ION, FLAVIN MONONUCLEOTIDE, Oxygen-insensitive NAD(P)H nitroreductase | | 著者 | Race, P.R, Lovering, A.L, Green, R.M, Ossor, A, White, S.A, Searle, P.F, Wrighton, C.J, Hyde, E.I. | | 登録日 | 2005-01-19 | | 公開日 | 2005-02-08 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structural and mechanistic studies of Escherichia coli nitroreductase with the antibiotic nitrofurazone. Reversed binding orientations in different redox states of the enzyme.
J.Biol.Chem., 280, 2005
|
|
1AVE
 
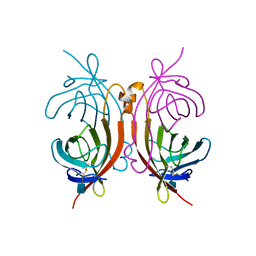 | | CRYSTAL STRUCTURE OF HEN EGG-WHITE APO-AVIDIN IN RELATION TO ITS THERMAL STABILITY PROPERTIES | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, AVIDIN | | 著者 | Pugliese, L, Coda, A, Malcovati, M, Bolognesi, M. | | 登録日 | 1993-03-05 | | 公開日 | 1994-01-31 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Crystal structure of apo-avidin from hen egg-white.
J.Mol.Biol., 235, 1994
|
|
1QGF
 
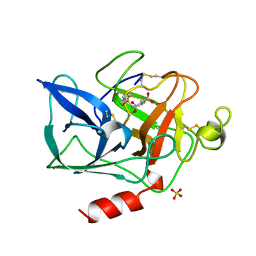 | | PORCINE PANCREATIC ELASTASE COMPLEXED WITH (3R, 4S)N-PARA-TOLUENESULPHONYL-3-ETHYL-4-(CARBOXYLIC ACID)PYRROLIDIN-2-ONE | | 分子名称: | (2S,3S)-3-FORMYL-2-({[(4-METHYLPHENYL)SULFONYL]AMINO}METHYL)PENTANOIC ACID, CALCIUM ION, ELASTASE, ... | | 著者 | Wilmouth, R.C, Kassamally, S, Westwood, N.J, Sheppard, R.J, Claridge, T.D.W, Wright, P.A, Pritchard, G.J, Schofield, C.J. | | 登録日 | 1999-04-27 | | 公開日 | 1999-12-29 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Mechanistic insights into the inhibition of serine proteases by monocyclic lactams.
Biochemistry, 38, 1999
|
|
1LR1
 
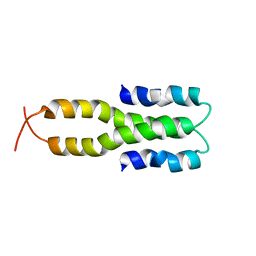 | | Solution Structure of the Oligomerization Domain of the Bacterial Chromatin-Structuring Protein H-NS | | 分子名称: | dna-binding protein h-ns | | 著者 | Esposito, D, Petrovic, A, Harris, R, Ono, S, Eccleston, J, Mbabaali, A, Haq, I, Higgins, C.F, Hinton, J.C.D, Driscoll, P.C, Ladbury, J.E. | | 登録日 | 2002-05-14 | | 公開日 | 2003-01-14 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | H-NS Oligomerization Domain Structure Reveals the Mechanism for High Order
Self-association of the Intact Protein
J.Mol.Biol., 324, 2002
|
|
1GID
 
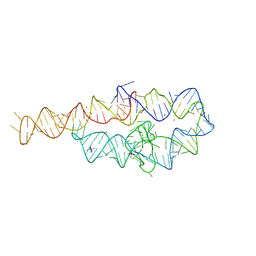 | | CRYSTAL STRUCTURE OF A GROUP I RIBOZYME DOMAIN: PRINCIPLES OF RNA PACKING | | 分子名称: | COBALT HEXAMMINE(III), MAGNESIUM ION, P4-P6 RNA RIBOZYME DOMAIN | | 著者 | Cate, J.H, Gooding, A.R, Podell, E, Zhou, K, Golden, B.L, Kundrot, C.E, Cech, T.R, Doudna, J.A. | | 登録日 | 1996-08-22 | | 公開日 | 1996-12-31 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Crystal structure of a group I ribozyme domain: principles of RNA packing.
Science, 273, 1996
|
|
1BET
 
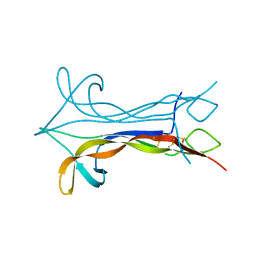 | | NEW PROTEIN FOLD REVEALED BY A 2.3 ANGSTROM RESOLUTION CRYSTAL STRUCTURE OF NERVE GROWTH FACTOR | | 分子名称: | BETA-NERVE GROWTH FACTOR | | 著者 | Mcdonald, N.Q, Lapatto, R, Murray-Rust, J, Gunning, J, Wlodawer, A, Blundell, T.L. | | 登録日 | 1993-04-08 | | 公開日 | 1994-05-31 | | 最終更新日 | 2024-06-05 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | New protein fold revealed by a 2.3-A resolution crystal structure of nerve growth factor.
Nature, 354, 1991
|
|
1ZY8
 
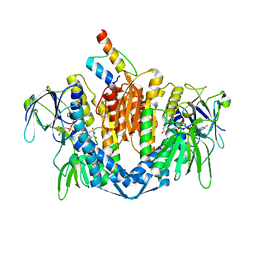 | | The crystal structure of dihydrolipoamide dehydrogenase and dihydrolipoamide dehydrogenase-binding protein (didomain) subcomplex of human pyruvate dehydrogenase complex. | | 分子名称: | Dihydrolipoyl dehydrogenase, mitochondrial, FLAVIN-ADENINE DINUCLEOTIDE, ... | | 著者 | Ciszak, E.M, Makal, A, Hong, Y.S, Vettaikkorumakankauv, A.K, Korotchkina, L.G, Patel, M.S. | | 登録日 | 2005-06-09 | | 公開日 | 2005-11-15 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.59 Å) | | 主引用文献 | How Dihydrolipoamide Dehydrogenase-binding Protein Binds Dihydrolipoamide Dehydrogenase in the Human Pyruvate Dehydrogenase Complex.
J.Biol.Chem., 281, 2006
|
|
1GN0
 
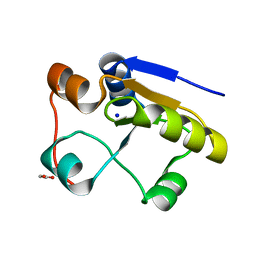 | | Escherichia coli GlpE sulfurtransferase soaked with KCN | | 分子名称: | 1,2-ETHANEDIOL, SODIUM ION, THIOSULFATE SULFURTRANSFERASE GLPE | | 著者 | Spallarossa, A, Donahue, J.T, Larson, T.J, Bolognesi, M, Bordo, D. | | 登録日 | 2001-10-01 | | 公開日 | 2001-11-29 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Escherichia Coli Glpe is a Prototype Sulfurtransferase for the Single-Domain Rhodanese Homology Superfamily
Structure, 9, 2001
|
|
1LMA
 
 | |
220L
 
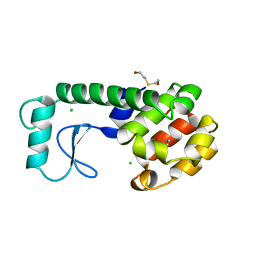 | | GENERATING LIGAND BINDING SITES IN T4 LYSOZYME USING DEFICIENCY-CREATING SUBSTITUTIONS | | 分子名称: | BENZENE, BETA-MERCAPTOETHANOL, CHLORIDE ION, ... | | 著者 | Baldwin, E.P, Baase, W.A, Zhang, X.-J, Feher, V, Matthews, B.W. | | 登録日 | 1997-06-25 | | 公開日 | 1998-03-18 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Generation of ligand binding sites in T4 lysozyme by deficiency-creating substitutions.
J.Mol.Biol., 277, 1998
|
|
1C61
 
 | |
1UWO
 
 | |
1C6H
 
 | |
1C6R
 
 | | CRYSTAL STRUCTURE OF REDUCED CYTOCHROME C6 FROM THE GREEN ALGAE SCENEDESMUS OBLIQUUS | | 分子名称: | CADMIUM ION, CYTOCHROME C6, PROTOPORPHYRIN IX CONTAINING FE | | 著者 | Schnackenberg, J, Than, M.E, Mann, K, Wiegand, G, Huber, R, Reuter, W. | | 登録日 | 1999-04-06 | | 公開日 | 2000-04-12 | | 最終更新日 | 2023-08-09 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Amino acid sequence, crystallization and structure determination of reduced and oxidized cytochrome c6 from the green alga Scenedesmus obliquus.
J.Mol.Biol., 290, 1999
|
|
1C60
 
 | |
1C62
 
 | |
1LTS
 
 | |
1C65
 
 | |
