4BJR
 
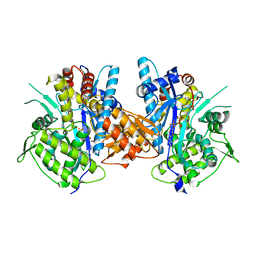 | | Crystal structure of the complex between Prokaryotic Ubiquitin-like Protein Pup and its Ligase PafA | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, PUP--PROTEIN LIGASE, ... | | 著者 | Barandun, J, Delley, C.L, Ban, N, Weber-Ban, E. | | 登録日 | 2013-04-19 | | 公開日 | 2013-05-01 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Crystal Structure of the Complex between Prokaryotic Ubiquitin-Like Protein Pup and its Ligase Pafa.
J.Am.Chem.Soc., 135, 2013
|
|
1HYO
 
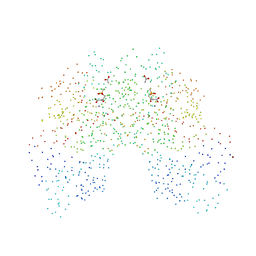 | | CRYSTAL STRUCTURE OF FUMARYLACETOACETATE HYDROLASE COMPLEXED WITH 4-(HYDROXYMETHYLPHOSPHINOYL)-3-OXO-BUTANOIC ACID | | 分子名称: | 4-[HYDROXY-[METHYL-PHOSPHINOYL]]-3-OXO-BUTANOIC ACID, ACETATE ION, CALCIUM ION, ... | | 著者 | Bateman, R.L, Bhanumoorthy, P, Witte, J.F, McClard, R.W, Grompe, M, Timm, D.E. | | 登録日 | 2001-01-21 | | 公開日 | 2001-02-14 | | 最終更新日 | 2023-08-09 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | Mechanistic inferences from the crystal structure of fumarylacetoacetate hydrolase with a bound phosphorus-based inhibitor.
J.Biol.Chem., 276, 2001
|
|
4C5P
 
 | | The structure of mycobacterium marinum arylamine n-acetyltransferase | | 分子名称: | ARYLAMINE N-ACETYLTRANSFERASE, AZIDE ION, GLYCEROL, ... | | 著者 | Abuhammad, A, Lowe, E.D, Garman, E.F, Sim, E. | | 登録日 | 2013-09-13 | | 公開日 | 2013-10-02 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (1.592 Å) | | 主引用文献 | Arylamine N-Acetyltransferases from Mycobacteria: Investigations of a Potential Target for Anti- Tubercular Therapy
Ph D Thesis
|
|
3VCA
 
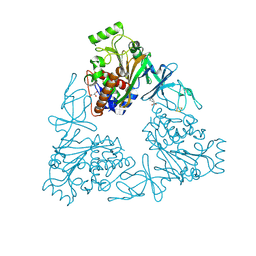 | | Quaternary Ammonium Oxidative Demethylation: X-ray Crystallographic, Resonance Raman and UV-visible Spectroscopic Analysis of a Rieske-type Demethylase | | 分子名称: | FE (III) ION, FE2/S2 (INORGANIC) CLUSTER, GLYCEROL, ... | | 著者 | Daughtry, K.D, Xiao, Y, Stoner-Ma, D, Cho, E, Orville, A.M, Liu, P, Allen, K.N. | | 登録日 | 2012-01-03 | | 公開日 | 2012-02-08 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (1.59 Å) | | 主引用文献 | Quaternary Ammonium Oxidative Demethylation: X-ray Crystallographic, Resonance Raman, and UV-Visible Spectroscopic Analysis of a Rieske-Type Demethylase.
J.Am.Chem.Soc., 134, 2012
|
|
2TRH
 
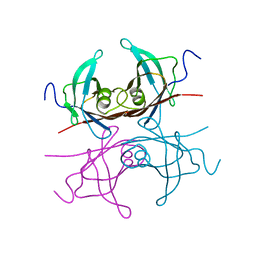 | |
2TRY
 
 | |
1IPB
 
 | | CRYSTAL STRUCTURE OF EUKARYOTIC INITIATION FACTOR 4E COMPLEXED WITH 7-METHYL GPPPA | | 分子名称: | EUKARYOTIC TRANSLATION INITIATION FACTOR 4E, P1-7-METHYLGUANOSINE-P3-ADENOSINE-5',5'-TRIPHOSPHATE | | 著者 | Tomoo, K, Shen, X, Okabe, K, Nozoe, Y, Fukuhara, S, Morino, S, Ishida, T, Taniguchi, T, Hasegawa, H, Terashima, A, Sasaki, M, Katsuya, Y, Kitamura, K, Miyoshi, H, Ishikawa, M, Miura, K. | | 登録日 | 2001-05-08 | | 公開日 | 2002-05-08 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal structures of 7-methylguanosine 5'-triphosphate (m(7)GTP)- and
P(1)-7-methylguanosine-P(3)-adenosine-5',5'-triphosphate (m(7)GpppA)-bound human full-length eukaryotic
initiation factor 4E: biological importance of the C-terminal flexible region
BIOCHEM.J., 362, 2002
|
|
1SPF
 
 | |
1E1Z
 
 | | Crystal structure of an Arylsulfatase A mutant C69S | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Arylsulfatase A, MAGNESIUM ION | | 著者 | von Buelow, R, Schmidt, B, Dierks, T, von Figura, K, Uson, I. | | 登録日 | 2000-05-12 | | 公開日 | 2001-05-10 | | 最終更新日 | 2024-11-13 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Crystal structure of an enzyme-substrate complex provides insight into the interaction between human arylsulfatase A and its substrates during catalysis.
J. Mol. Biol., 305, 2001
|
|
1E3C
 
 | | Crystal structure of an Arylsulfatase A mutant C69S soaked in synthetic substrate | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Arylsulfatase A, MAGNESIUM ION | | 著者 | von Buelow, R, Schmidt, B, Dierks, T, von Figura, K, Uson, I. | | 登録日 | 2000-06-13 | | 公開日 | 2001-03-05 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (2.65 Å) | | 主引用文献 | Crystal structure of an enzyme-substrate complex provides insight into the interaction between human arylsulfatase A and its substrates during catalysis.
J. Mol. Biol., 305, 2001
|
|
1BK5
 
 | | KARYOPHERIN ALPHA FROM SACCHAROMYCES CEREVISIAE | | 分子名称: | COBALT (II) ION, KARYOPHERIN ALPHA | | 著者 | Conti, E, Uy, M, Leighton, L, Blobel, G, Kuriyan, J. | | 登録日 | 1998-07-14 | | 公開日 | 1999-01-06 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Crystallographic analysis of the recognition of a nuclear localization signal by the nuclear import factor karyopherin alpha.
Cell(Cambridge,Mass.), 94, 1998
|
|
1BZD
 
 | |
1BZE
 
 | |
2L6E
 
 | | NMR Structure of the monomeric mutant C-terminal domain of HIV-1 Capsid in complex with stapled peptide Inhibitor | | 分子名称: | Capsid protein p24, NYAD-13 stapled peptide inhibitor | | 著者 | Bhattacharya, S, Zhang, H, Debnath, A.K, Cowburn, D. | | 登録日 | 2010-11-18 | | 公開日 | 2010-12-29 | | 最終更新日 | 2024-10-16 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of a hydrocarbon stapled peptide inhibitor in complex with monomeric C-terminal domain of HIV-1 capsid.
J.Biol.Chem., 283, 2008
|
|
1VA4
 
 | | Pseudomonas fluorescens aryl esterase | | 分子名称: | Arylesterase, GLYCEROL | | 著者 | Cheeseman, J.D, Tocilj, A, Park, S, Schrag, J.D, Kazlauskas, R.J. | | 登録日 | 2004-02-11 | | 公開日 | 2004-07-06 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.804 Å) | | 主引用文献 | Structure of an aryl esterase from Pseudomonas fluorescens.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1W4T
 
 | | X-ray crystallographic structure of Pseudomonas aeruginosa arylamine N-acetyltransferase | | 分子名称: | Arylamine N-acetyltransferase, SULFATE ION | | 著者 | Westwood, I.M, Holton, S.J, Rodrigues-Lima, F, Dupret, J.-M, Bhakta, S, Noble, M.E, Sim, E. | | 登録日 | 2004-07-29 | | 公開日 | 2005-08-03 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Expression, purification, characterization and structure of Pseudomonas aeruginosa arylamine N-acetyltransferase.
Biochem. J., 385, 2005
|
|
1XPL
 
 | | Crystal Structure of Staphylococcus aureus HMG-COA Synthase with Acetoacetyl-COA and Acetylated Cysteine | | 分子名称: | 3-hydroxy-3-methylglutaryl CoA synthase, ACETOACETYL-COENZYME A, SULFATE ION | | 著者 | Theisen, M.J, Misra, I, Saadat, D, Campobasso, N, Miziorko, H.M, Harrison, D.H.T. | | 登録日 | 2004-10-08 | | 公開日 | 2004-11-02 | | 最終更新日 | 2024-11-20 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | 3-hydroxy-3-methylglutaryl-CoA synthase intermediate complex observed in "real-time"
Proc.Natl.Acad.Sci.USA, 47, 2004
|
|
2LKB
 
 | | Evolutionary diversification of Mesobuthus alpha-scorpion toxins affecting sodium channels | | 分子名称: | Neurotoxin MeuNaTx-5 | | 著者 | Zhu, S, Peigneur, S, Gao, B, Lu, X, Cao, C, Tytgat, J. | | 登録日 | 2011-10-09 | | 公開日 | 2011-11-02 | | 最終更新日 | 2024-10-30 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Evolutionary diversification of Mesobuthus {alpha}-scorpion toxins affecting sodium channels
Mol.Cell Proteomics, 2011
|
|
1QBK
 
 | |
1E2S
 
 | | Crystal structure of an Arylsulfatase A mutant C69A | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Arylsulfatase A, MAGNESIUM ION, ... | | 著者 | von Buelow, R, Schmidt, B, Dierks, T, von Figura, K, Uson, I. | | 登録日 | 2000-05-24 | | 公開日 | 2000-12-06 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | Crystal structure of an enzyme-substrate complex provides insight into the interaction between human arylsulfatase A and its substrates during catalysis.
J. Mol. Biol., 305, 2001
|
|
8DID
 
 | |
8DIC
 
 | |
8DIB
 
 | |
8DIF
 
 | |
8DIH
 
 | | Virtual screening for novel SARS-CoV-2 main protease non-covalent and covalent inhibitors | | 分子名称: | (1P,1'R)-1-(isoquinolin-4-yl)-2',3'-dihydrospiro[imidazolidine-4,1'-indene]-2,5-dione, 3C-like proteinase nsp5 | | 著者 | Singh, I, Shoichet, B.K. | | 登録日 | 2022-06-29 | | 公開日 | 2023-06-28 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.12 Å) | | 主引用文献 | Large library docking for novel SARS-CoV-2 main protease non-covalent and covalent inhibitors.
Protein Sci., 32, 2023
|
|
