4BDQ
 
 | | Crystal structure of the GluK2 R775A LBD dimer in complex with glutamate | | 分子名称: | GLUTAMATE RECEPTOR, IONOTROPIC KAINATE 2, GLUTAMIC ACID, ... | | 著者 | Nayeem, N, Mayans, O, Green, T. | | 登録日 | 2012-10-05 | | 公開日 | 2013-04-10 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Correlating Efficacy and Desensitization with Gluk2 Ligand-Binding Domain Movements.
Open Biol., 3, 2013
|
|
4BDL
 
 | | Crystal structure of the GluK2 K531A LBD dimer in complex with glutamate | | 分子名称: | GLUTAMATE RECEPTOR, IONOTROPIC KAINATE 2, GLUTAMIC ACID, ... | | 著者 | Nayeem, N, Mayans, O, Green, T. | | 登録日 | 2012-10-05 | | 公開日 | 2013-04-10 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Correlating Efficacy and Desensitization with Gluk2 Ligand-Binding Domain Movements.
Open Biol., 3, 2013
|
|
4BDO
 
 | | Crystal structure of the GluK2 K531A-T779G LBD dimer in complex with kainate | | 分子名称: | 3-(CARBOXYMETHYL)-4-ISOPROPENYLPROLINE, GLUTAMATE RECEPTOR, IONOTROPIC KAINATE 2, ... | | 著者 | Nayeem, N, Mayans, O, Green, T. | | 登録日 | 2012-10-05 | | 公開日 | 2013-04-10 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.55 Å) | | 主引用文献 | Correlating Efficacy and Desensitization with Gluk2 Ligand-Binding Domain Movements.
Open Biol., 3, 2013
|
|
4BDM
 
 | |
4BDN
 
 | | Crystal structure of the GluK2 K531A-T779G LBD dimer in complex with glutamate | | 分子名称: | GLUTAMATE RECEPTOR, IONOTROPIC KAINATE 2, GLUTAMIC ACID, ... | | 著者 | Nayeem, N, Mayans, O, Green, T. | | 登録日 | 2012-10-05 | | 公開日 | 2013-04-10 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Correlating Efficacy and Desensitization with Gluk2 Ligand-Binding Domain Movements.
Open Biol., 3, 2013
|
|
4BDR
 
 | | Crystal structure of the GluK2 R775A LBD dimer in complex with kainate | | 分子名称: | 3-(CARBOXYMETHYL)-4-ISOPROPENYLPROLINE, CHLORIDE ION, GLUTAMATE RECEPTOR, ... | | 著者 | Nayeem, N, Mayans, O, Green, T. | | 登録日 | 2012-10-05 | | 公開日 | 2013-04-10 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Correlating Efficacy and Desensitization with Gluk2 Ligand-Binding Domain Movements.
Open Biol., 3, 2013
|
|
4IGR
 
 | | Crystal structure of the kainate receptor GluK3 ligand-binding domain in complex with the agonist ZA302 | | 分子名称: | (4R)-4-{3-[hydroxy(methyl)amino]-3-oxopropyl}-L-glutamic acid, CHLORIDE ION, Glutamate receptor, ... | | 著者 | Larsen, A.P, Venskutonyte, R, Gajhede, M, Kastrup, J.S, Frydenvang, K. | | 登録日 | 2012-12-18 | | 公開日 | 2013-03-06 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.65 Å) | | 主引用文献 | Chemoenzymatic synthesis of new 2,4-syn-functionalized (S)-glutamate analogues and structure-activity relationship studies at ionotropic glutamate receptors and excitatory amino acid transporters.
J.Med.Chem., 56, 2013
|
|
4IGT
 
 | | Crystal structure of the GluA2 ligand-binding domain (S1S2J) in complex with the agonist ZA302 at 1.24A resolution | | 分子名称: | (4R)-4-{3-[hydroxy(methyl)amino]-3-oxopropyl}-L-glutamic acid, GLYCEROL, Glutamate receptor 2, ... | | 著者 | Larsen, A.P, Venskutonyte, R, Gajhede, M, Kastrup, J.S, Frydenvang, K. | | 登録日 | 2012-12-18 | | 公開日 | 2013-03-06 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.24 Å) | | 主引用文献 | Chemoenzymatic synthesis of new 2,4-syn-functionalized (S)-glutamate analogues and structure-activity relationship studies at ionotropic glutamate receptors and excitatory amino acid transporters.
J.Med.Chem., 56, 2013
|
|
4IO5
 
 | |
4IO2
 
 | |
4IO7
 
 | |
4IO3
 
 | |
4IO6
 
 | |
4IO4
 
 | | Crystal Structure of the AvGluR1 ligand binding domain complex with serine at 1.94 Angstrom resolution | | 分子名称: | AvGluR1 ligand binding domain, CHLORIDE ION, GLYCEROL, ... | | 著者 | Lomash, S, Chittori, S, Mayer, M.L. | | 登録日 | 2013-01-07 | | 公開日 | 2013-02-20 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.941 Å) | | 主引用文献 | Anions Mediate Ligand Binding in Adineta vaga Glutamate Receptor Ion Channels.
Structure, 21, 2013
|
|
4ISU
 
 | | Crystal structure of the GluA2 ligand-binding domain (S1S2J) in complex with the antagonist (2R)-IKM-159 at 2.3A resolution. | | 分子名称: | (4aS,5aR,6R,8aS,8bS)-5a-(carboxymethyl)-8-oxo-2,4a,5a,6,7,8,8a,8b-octahydro-1H-pyrrolo[3',4':4,5]furo[3,2-b]pyridine-6-carboxylic acid, CHLORIDE ION, Glutamate receptor 2, ... | | 著者 | Juknaite, L, Frydenvang, K, Kastrup, J.S. | | 登録日 | 2013-01-17 | | 公開日 | 2013-03-20 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Studies on an (S)-2-amino-3-(3-hydroxy-5-methyl-4-isoxazolyl)propionic acid (AMPA) receptor antagonist IKM-159: asymmetric synthesis, neuroactivity, and structural characterization.
J.Med.Chem., 56, 2013
|
|
4IY5
 
 | | Crystal structure of the glua2 ligand-binding domain (S1S2J-L483Y-N754S) in complex with glutamate and CX516 at 2.0 A resolution | | 分子名称: | CHLORIDE ION, GLUTAMIC ACID, GLYCEROL, ... | | 著者 | Krintel, C, Frydenvang, K, Harpsoe, K, Gajhede, M, Kastrup, J.S. | | 登録日 | 2013-01-28 | | 公開日 | 2013-10-09 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural analysis of the positive AMPA receptor modulators CX516 and Me-CX516 in complex with the GluA2 ligand-binding domain
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4IY6
 
 | | Crystal structure of the GLUA2 ligand-binding domain (S1S2J-L483Y-N754S) in complex with glutamate and ME-CX516 at 1.72 A resolution | | 分子名称: | GLUTAMIC ACID, GLYCEROL, Glutamate receptor 2, ... | | 著者 | Krintel, C, Frydenvang, K, Harpsoe, K, Gajhede, M, Kastrup, J.S. | | 登録日 | 2013-01-28 | | 公開日 | 2013-10-09 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.72 Å) | | 主引用文献 | Structural analysis of the positive AMPA receptor modulators CX516 and Me-CX516 in complex with the GluA2 ligand-binding domain
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4JWY
 
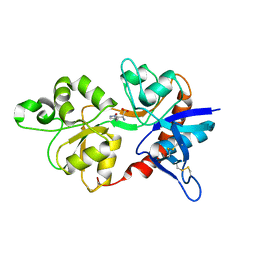 | | GluN2D ligand-binding core in complex with propyl-NHP5G | | 分子名称: | (2R)-amino(1-hydroxy-4-propyl-1H-pyrazol-5-yl)ethanoic acid, Glutamate receptor ionotropic, NMDA 2D | | 著者 | Hansen, K.B, Tajima, N, Risgaard, R, Perszyk, R.E, Jorgensen, L, Vance, K.M, Ogden, K.K, Clausen, R.P, Furukawa, H, Traynelis, S.F. | | 登録日 | 2013-03-27 | | 公開日 | 2013-05-29 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural determinants of agonist efficacy at the glutamate binding site of N-methyl-d-aspartate receptors.
Mol.Pharmacol., 84, 2013
|
|
4JWX
 
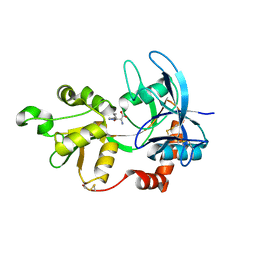 | | GluN2A ligand-binding core in complex with propyl-NHP5G | | 分子名称: | (2R)-amino(1-hydroxy-4-propyl-1H-pyrazol-5-yl)ethanoic acid, GluN2A | | 著者 | Hansen, K.B, Tajima, N, Risgaard, R, Perszyk, R.E, Jorgensen, L, Vance, K.M, Ogden, K.K, Clausen, R.P, Furukawa, H, Traynelis, S.F. | | 登録日 | 2013-03-27 | | 公開日 | 2013-05-29 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Structural determinants of agonist efficacy at the glutamate binding site of N-methyl-d-aspartate receptors.
Mol.Pharmacol., 84, 2013
|
|
4KCD
 
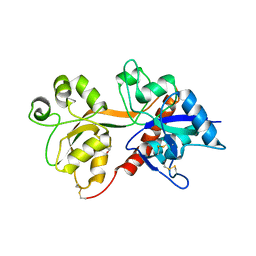 | | Crystal Structure of the NMDA Receptor GluN3A Ligand Binding Domain Apo State | | 分子名称: | GLYCEROL, Glutamate receptor ionotropic, NMDA 3A | | 著者 | Yao, Y, Lau, A.Y, Mayer, M.L. | | 登録日 | 2013-04-24 | | 公開日 | 2013-07-31 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.68 Å) | | 主引用文献 | Conformational Analysis of NMDA Receptor GluN1, GluN2, and GluN3 Ligand-Binding Domains Reveals Subtype-Specific Characteristics.
Structure, 21, 2013
|
|
4KCC
 
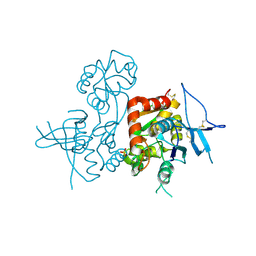 | | Crystal Structure of the NMDA Receptor GluN1 Ligand Binding Domain Apo State | | 分子名称: | Glutamate receptor ionotropic, NMDA 1, PHOSPHATE ION | | 著者 | Berger, A.J, Lau, A.Y, Mayer, M.L. | | 登録日 | 2013-04-24 | | 公開日 | 2013-07-31 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.894 Å) | | 主引用文献 | Conformational Analysis of NMDA Receptor GluN1, GluN2, and GluN3 Ligand-Binding Domains Reveals Subtype-Specific Characteristics.
Structure, 21, 2013
|
|
4KFQ
 
 | | Crystal structure of the NMDA receptor GluN1 ligand binding domain in complex with 1-thioxo-1,2-dihydro-[1,2,4]triazolo[4,3-a]quinoxalin-4(5H)-one | | 分子名称: | 1-sulfanyl[1,2,4]triazolo[4,3-a]quinoxalin-4(5H)-one, GLYCEROL, Glutamate receptor ionotropic, ... | | 著者 | Steffensen, T.B, Tabrizi, F.M, Gajhede, M, Kastrup, J.S. | | 登録日 | 2013-04-27 | | 公開日 | 2013-10-09 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Crystal structure and pharmacological characterization of a novel N-methyl-D-aspartate (NMDA) receptor antagonist at the GluN1 glycine binding site.
J.Biol.Chem., 288, 2013
|
|
4L17
 
 | | GluA2-L483Y-A665C ligand-binding domain in complex with the antagonist DNQX | | 分子名称: | 6,7-DINITROQUINOXALINE-2,3-DIONE, Glutamate receptor 2, SULFATE ION | | 著者 | Lau, A.Y, Blachowicz, L, Roux, B. | | 登録日 | 2013-06-02 | | 公開日 | 2013-08-14 | | 最終更新日 | 2017-08-02 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | A conformational intermediate in glutamate receptor activation.
Neuron, 79, 2013
|
|
4LZ5
 
 | |
4LZ8
 
 | |
