8G2U
 
 | | Time-resolved cryo-EM study of the 70S recycling by the HflX:control-apo-70S at 900ms | | 分子名称: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | 著者 | Bhattacharjee, S, Brown, P.Z, Frank, J. | | 登録日 | 2023-02-06 | | 公開日 | 2023-12-06 | | 最終更新日 | 2024-02-14 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Time resolution in cryo-EM using a PDMS-based microfluidic chip assembly and its application to the study of HflX-mediated ribosome recycling.
Cell, 187, 2024
|
|
6X7K
 
 | | Cryo-EM structure of an Escherichia coli coupled transcription-translation complex B3 (TTC-B3) containing an mRNA with a 24 nt long spacer, transcription factors NusA and NusG, and fMet-tRNAs at P-site and E-site | | 分子名称: | 16S rRNA, 23S rRNA, 30S ribosomal protein S1, ... | | 著者 | Molodtsov, V, Ebright, R.H, Wang, C, Su, M. | | 登録日 | 2020-05-30 | | 公開日 | 2020-09-02 | | 最終更新日 | 2020-09-23 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Structural basis of transcription-translation coupling.
Science, 369, 2020
|
|
6X6T
 
 | | Cryo-EM structure of an Escherichia coli coupled transcription-translation complex B1 (TTC-B1) containing an mRNA with a 24 nt long spacer, transcription factors NusA and NusG, and fMet-tRNAs at P-site and E-site | | 分子名称: | 16S rRNA, 23S rRNA, 30S ribosomal protein S1, ... | | 著者 | Molodtsov, V, Ebright, R.H, Wang, C, Su, M. | | 登録日 | 2020-05-29 | | 公開日 | 2020-09-02 | | 最終更新日 | 2020-09-23 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Structural basis of transcription-translation coupling.
Science, 369, 2020
|
|
8G34
 
 | | Time-resolved cryo-EM study of the 70S recycling by the HflX:1st intermediate | | 分子名称: | 16S, 23S, 30S ribosomal protein S10, ... | | 著者 | Bhattacharjee, S, Brown, P.Z, Frank, J. | | 登録日 | 2023-02-06 | | 公開日 | 2023-12-06 | | 最終更新日 | 2024-02-14 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Time resolution in cryo-EM using a PDMS-based microfluidic chip assembly and its application to the study of HflX-mediated ribosome recycling.
Cell, 187, 2024
|
|
8G38
 
 | | Time-resolved cryo-EM study of the 70S recycling by the HflX:3rd Intermediate | | 分子名称: | 16S, 23S, 30S ribosomal protein S10, ... | | 著者 | Bhattacharjee, S, Brown, P.Z, Frank, J. | | 登録日 | 2023-02-07 | | 公開日 | 2023-12-06 | | 最終更新日 | 2024-02-14 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Time resolution in cryo-EM using a PDMS-based microfluidic chip assembly and its application to the study of HflX-mediated ribosome recycling.
Cell, 187, 2024
|
|
8G31
 
 | | Time-resolved cryo-EM study of the 70S recycling by the HflX:2nd Intermediate | | 分子名称: | 16S, 23S, 30S ribosomal protein S10, ... | | 著者 | Bhattacharjee, S, Brown, P.Z, Frank, J. | | 登録日 | 2023-02-06 | | 公開日 | 2023-12-06 | | 最終更新日 | 2024-02-14 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Time resolution in cryo-EM using a PDMS-based microfluidic chip assembly and its application to the study of HflX-mediated ribosome recycling.
Cell, 187, 2024
|
|
6ZTJ
 
 | | E. coli 70S-RNAP expressome complex in NusG-coupled state (38 nt intervening mRNA) | | 分子名称: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S1, ... | | 著者 | Webster, M.W, Takacs, M, Weixlbaumer, A. | | 登録日 | 2020-07-20 | | 公開日 | 2020-09-16 | | 最終更新日 | 2024-04-24 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Structural basis of transcription-translation coupling and collision in bacteria.
Science, 369, 2020
|
|
6X7F
 
 | | Cryo-EM structure of an Escherichia coli coupled transcription-translation complex B2 (TTC-B2) containing an mRNA with a 24 nt long spacer, transcription factors NusA and NusG, and fMet-tRNAs at P-site and E-site | | 分子名称: | 16S rRNA, 23S rRNA, 30S ribosomal protein S1, ... | | 著者 | Molodtsov, V, Ebright, R.H, Wang, C, Su, M. | | 登録日 | 2020-05-29 | | 公開日 | 2020-09-02 | | 最終更新日 | 2020-09-23 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Structural basis of transcription-translation coupling.
Science, 369, 2020
|
|
6ZTL
 
 | | E. coli 70S-RNAP expressome complex in collided state bound to NusG | | 分子名称: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | 著者 | Webster, M.W, Takacs, M, Weixlbaumer, A. | | 登録日 | 2020-07-20 | | 公開日 | 2020-09-16 | | 最終更新日 | 2024-04-24 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Structural basis of transcription-translation coupling and collision in bacteria.
Science, 369, 2020
|
|
6VYS
 
 | | Escherichia coli transcription-translation complex A1 (TTC-A1) containing a 21 nt long mRNA spacer, NusG, and fMet-tRNAs at E-site and P-site | | 分子名称: | 16S rRNA, 23S rRNA, 30S ribosomal protein S1, ... | | 著者 | Molodtsov, V, Wang, C, Su, M, Ebright, R.H. | | 登録日 | 2020-02-27 | | 公開日 | 2020-09-02 | | 最終更新日 | 2020-09-23 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Structural basis of transcription-translation coupling.
Science, 369, 2020
|
|
6VYQ
 
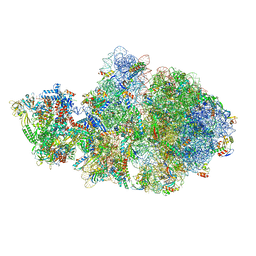 | | Escherichia coli transcription-translation complex A1 (TTC-A1) containing an 15 nt long mRNA spacer, NusG, and fMet-tRNAs at E-site and P-site | | 分子名称: | 16S rRNA, 23S rRNA, 30S ribosomal protein S1, ... | | 著者 | Molodtsov, V, Wang, C, Su, M, Ebright, R.H. | | 登録日 | 2020-02-27 | | 公開日 | 2020-09-02 | | 最終更新日 | 2020-09-23 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Structural basis of transcription-translation coupling.
Science, 369, 2020
|
|
6GOV
 
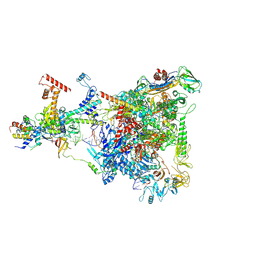 | | Structure of THE RNA POLYMERASE LAMBDA-BASED ANTITERMINATION COMPLEX | | 分子名称: | 30S ribosomal protein S10, Antitermination protein N, DNA (I), ... | | 著者 | Loll, B, Krupp, F, Said, N, Huang, Y, Buerger, J, Mielke, T, Spahn, C.M.T, Wahl, M.C. | | 登録日 | 2018-06-04 | | 公開日 | 2019-02-13 | | 最終更新日 | 2019-12-18 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Structural Basis for the Action of an All-Purpose Transcription Anti-termination Factor.
Mol.Cell, 74, 2019
|
|
6XDQ
 
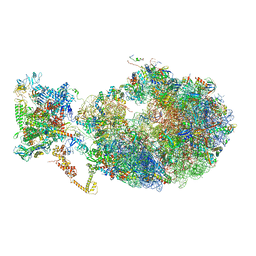 | | Cryo-EM structure of an Escherichia coli coupled transcription-translation complex B3 (TTC-B3) containing an mRNA with a 30 nt long spacer, transcription factors NusA and NusG, and fMet-tRNAs at P-site and E-site | | 分子名称: | 16S rRNA, 23S rRNA, 30S ribosomal protein S1, ... | | 著者 | Molodtsov, V, Ebright, R.H, Wang, C, Su, M. | | 登録日 | 2020-06-11 | | 公開日 | 2020-09-02 | | 最終更新日 | 2020-09-23 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Structural basis of transcription-translation coupling.
Science, 369, 2020
|
|
6DUQ
 
 | | Structure of a Rho-NusG KOW domain complex | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, MAGNESIUM ION, ... | | 著者 | Berger, J.M, Lawson, M.R. | | 登録日 | 2018-06-21 | | 公開日 | 2018-09-05 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (3.7 Å) | | 主引用文献 | Mechanism for the Regulated Control of Bacterial Transcription Termination by a Universal Adaptor Protein.
Mol. Cell, 71, 2018
|
|
6VU3
 
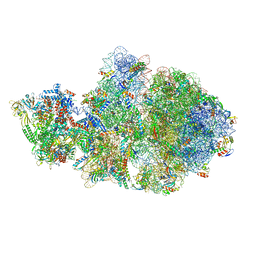 | | Cryo-EM structure of Escherichia coli transcription-translation complex A (TTC-A) containing mRNA with a 12 nt long spacer | | 分子名称: | 16S rRNA, 23S rRNA, 30S ribosomal protein S1, ... | | 著者 | Molodtsov, V, Wang, C, Su, M, Ebright, R. | | 登録日 | 2020-02-14 | | 公開日 | 2020-09-02 | | 最終更新日 | 2020-09-23 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Structural basis of transcription-translation coupling.
Science, 369, 2020
|
|
6C6U
 
 | | CryoEM structure of E.coli RNA polymerase elongation complex bound with NusG | | 分子名称: | DNA (29-MER), DNA-DIRECTED RNA POLYMERASE BETA', DNA-directed RNA polymerase subunit alpha, ... | | 著者 | Kang, J.Y, Artsimovitch, I, Landick, R, Darst, S.A. | | 登録日 | 2018-01-19 | | 公開日 | 2018-07-25 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Structural Basis for Transcript Elongation Control by NusG Family Universal Regulators.
Cell, 173, 2018
|
|
8E79
 
 | | Mycobacterium tuberculosis RNAP paused elongation complex with Escherichia coli NusG transcription factor | | 分子名称: | DNA (54-MER), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | 著者 | Delbeau, M, Darst, S.A, Campbell, E.A. | | 登録日 | 2022-08-23 | | 公開日 | 2023-03-22 | | 最終更新日 | 2023-10-11 | | 実験手法 | ELECTRON MICROSCOPY (3.71 Å) | | 主引用文献 | Structural and functional basis of the universal transcription factor NusG pro-pausing activity in Mycobacterium tuberculosis.
Mol.Cell, 83, 2023
|
|
6TQO
 
 | | rrn anti-termination complex | | 分子名称: | 30S ribosomal protein S10, 30S ribosomal protein S4, DNA-directed RNA polymerase subunit alpha, ... | | 著者 | Huang, Y.H, Wahl, M.C, Loll, B, Hilal, T, Said, N. | | 登録日 | 2019-12-17 | | 公開日 | 2020-08-05 | | 最終更新日 | 2024-05-15 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Structure-Based Mechanisms of a Molecular RNA Polymerase/Chaperone Machine Required for Ribosome Biosynthesis.
Mol.Cell, 79, 2020
|
|
7PY8
 
 | | CryoEM structure of E.coli RNA polymerase elongation complex bound to NusG (NusG-EC in less-swiveled conformation) | | 分子名称: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | 著者 | Zhu, C, Guo, X, Weixlbaumer, A. | | 登録日 | 2021-10-09 | | 公開日 | 2022-03-23 | | 最終更新日 | 2024-07-17 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Transcription factors modulate RNA polymerase conformational equilibrium.
Nat Commun, 13, 2022
|
|
7PY1
 
 | | CryoEM structure of E.coli RNA polymerase elongation complex bound to NusG (the consensus NusG-EC) | | 分子名称: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | 著者 | Zhu, C, Guo, X, Weixlbaumer, A. | | 登録日 | 2021-10-08 | | 公開日 | 2022-03-23 | | 最終更新日 | 2024-07-17 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Transcription factors modulate RNA polymerase conformational equilibrium.
Nat Commun, 13, 2022
|
|
6TQN
 
 | | rrn anti-termination complex without S4 | | 分子名称: | 30S ribosomal protein S10, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | 著者 | Huang, Y.H, Wahl, M.C, Loll, B, Hilal, T, Said, N. | | 登録日 | 2019-12-17 | | 公開日 | 2020-08-05 | | 最終更新日 | 2024-05-15 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Structure-Based Mechanisms of a Molecular RNA Polymerase/Chaperone Machine Required for Ribosome Biosynthesis.
Mol.Cell, 79, 2020
|
|
6VYR
 
 | | Escherichia coli transcription-translation complex A1 (TTC-A1) containing an 18 nt long mRNA spacer, NusG, and fMet-tRNAs at E-site and P-site | | 分子名称: | 16S rRNA, 23S rRNA, 30S ribosomal protein S1, ... | | 著者 | Molodtsov, V, Wang, C, Su, M, Ebright, R.H. | | 登録日 | 2020-02-27 | | 公開日 | 2020-09-02 | | 最終更新日 | 2020-09-23 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Structural basis of transcription-translation coupling.
Science, 369, 2020
|
|
6ZTN
 
 | | E. coli 70S-RNAP expressome complex in NusG-coupled state (42 nt intervening mRNA) | | 分子名称: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | 著者 | Webster, M.W, Takacs, M, Weixlbaumer, A. | | 登録日 | 2020-07-20 | | 公開日 | 2020-09-16 | | 最終更新日 | 2024-04-24 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Structural basis of transcription-translation coupling and collision in bacteria.
Science, 369, 2020
|
|
7PY5
 
 | | CryoEM structure of E.coli RNA polymerase elongation complex bound to NusA and NusG (the consensus NusA-NusG-EC) | | 分子名称: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | 著者 | Zhu, C, Guo, X, Weixlbaumer, A. | | 登録日 | 2021-10-09 | | 公開日 | 2022-03-23 | | 最終更新日 | 2024-07-17 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Transcription factors modulate RNA polymerase conformational equilibrium.
Nat Commun, 13, 2022
|
|
6Z9P
 
 | | Transcription termination intermediate complex 1 | | 分子名称: | 2'-DEOXYGUANOSINE-5'-MONOPHOSPHATE, ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, ... | | 著者 | Said, N, Hilal, T, Loll, B, Wahl, M.C. | | 登録日 | 2020-06-04 | | 公開日 | 2020-11-04 | | 最終更新日 | 2021-02-03 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Steps toward translocation-independent RNA polymerase inactivation by terminator ATPase rho.
Science, 371, 2021
|
|
