5RLR
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 helicase in complex with Z822382694 | | 分子名称: | (1R)-2-(methylsulfonyl)-1-phenylethan-1-ol, Helicase, PHOSPHATE ION, ... | | 著者 | Newman, J.A, Yosaatmadja, Y, Douangamath, A, Aimon, A, Powell, A.J, Dias, A, Fearon, D, Dunnett, L, Brandao-Neto, J, Krojer, T, Skyner, R, Gorrie-Stone, T, Thompson, W, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | 登録日 | 2020-09-16 | | 公開日 | 2020-09-30 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (2.32 Å) | | 主引用文献 | Structure, mechanism and crystallographic fragment screening of the SARS-CoV-2 NSP13 helicase.
Nat Commun, 12, 2021
|
|
8Q71
 
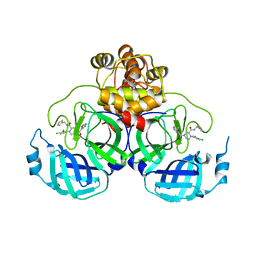 | | Crystal structure of SARS-CoV-2 main protease (MPro) in complex with the inhibitor GC-67 | | 分子名称: | (2~{S})-1-(3,4-dichlorophenyl)-4-(4-methoxypyridin-3-yl)carbonyl-~{N}-(thiophen-2-ylmethyl)piperazine-2-carboxamide, 3C-like proteinase nsp5 | | 著者 | Strater, N, Muller, C.E, Sylvester, K, Weisse, R.H, Useini, A, Gao, S, Song, L, Liu, Z, Zhan, P. | | 登録日 | 2023-08-15 | | 公開日 | 2023-12-06 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.322 Å) | | 主引用文献 | Design, Synthesis, and Biological Evaluation of Trisubstituted Piperazine Derivatives as Noncovalent Severe Acute Respiratory Syndrome Coronavirus 2 Main Protease Inhibitors with Improved Antiviral Activity and Favorable Druggability.
J.Med.Chem., 66, 2023
|
|
7NT3
 
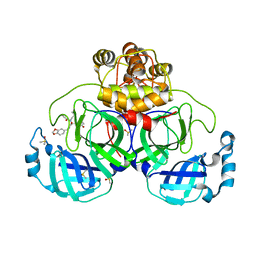 | | Crystal structure of SARS CoV2 main protease in complex with FSCU015 | | 分子名称: | 3C-like proteinase, DIMETHYL SULFOXIDE, ~{N}-[(1~{S})-2-(1,3-benzodioxol-5-ylmethylamino)-1-(3-hydroxyphenyl)-2-oxidanylidene-ethyl]-~{N}-propyl-prop-2-enamide | | 著者 | Oerlemans, R, Eris, D, Wang, M, Sharpe, M, Domling, A, Groves, M.R. | | 登録日 | 2021-03-08 | | 公開日 | 2021-06-16 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.325 Å) | | 主引用文献 | Combining High-Throughput Synthesis and High-Throughput Protein Crystallography for Accelerated Hit Identification.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
5S70
 
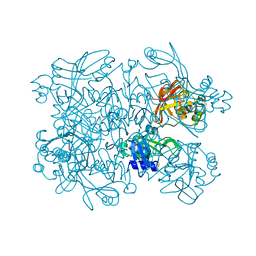 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 NendoU in complex with EN300-181428 | | 分子名称: | (5R)-2-methyl-4,5,6,7-tetrahydro-1H-benzimidazol-5-amine, CITRIC ACID, Uridylate-specific endoribonuclease | | 著者 | Godoy, A.S, Douangamath, A, Nakamura, A.M, Dias, A, Krojer, T, Noske, G.D, Gawiljuk, V.O, Fernandes, R.S, Fairhead, M, Powell, A, Dunnet, L, Aimon, A, Fearon, D, Brandao-Neto, J, Skyner, R, von Delft, F, Oliva, G. | | 登録日 | 2020-11-13 | | 公開日 | 2020-11-25 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (2.327 Å) | | 主引用文献 | Allosteric regulation and crystallographic fragment screening of SARS-CoV-2 NSP15 endoribonuclease.
Nucleic Acids Res., 2023
|
|
9EUN
 
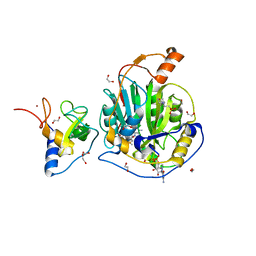 | | SARS-CoV-2 nsp10-16 methyltransferase in complex with SAM and m7GTP | | 分子名称: | 1,2-ETHANEDIOL, 2'-O-methyltransferase nsp16, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | 著者 | Kremling, V, Sprenger, J, Oberthuer, D, Scheer, T.E.S. | | 登録日 | 2024-03-27 | | 公開日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (2.33 Å) | | 主引用文献 | SARS-CoV-2 methyltransferase nsp10-16 in complex with natural and drug-like purine analogs for guiding structure-based drug development
To Be Published
|
|
8UD2
 
 | |
9EMV
 
 | | SARS-CoV-2 nsp10-16 methyltransferase in complex with Sangivamycin and m7GpppA (Cap0-analog)/m7GpppAm (Cap1-analog) | | 分子名称: | 1,2-ETHANEDIOL, 2'-O-methyltransferase nsp16, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | 著者 | Kremling, V, Sprenger, J, Oberthuer, D, Scheer, T.E.S. | | 登録日 | 2024-03-11 | | 公開日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.34 Å) | | 主引用文献 | SARS-CoV-2 methyltransferase nsp10-16 in complex with natural and drug-like purine analogs for guiding structure-based drug development
To Be Published
|
|
7X6K
 
 | | SARS-CoV-2 3CL protease (3CLpro) in complex with compound 3w | | 分子名称: | 1H-indole-2-carbaldehyde, 3C-like proteinase | | 著者 | Su, H, Nie, T, Xie, H, Li, Z.W, Li, M.J, Xu, Y. | | 登録日 | 2022-03-07 | | 公開日 | 2022-07-06 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.34 Å) | | 主引用文献 | Small-Molecule Thioesters as SARS-CoV-2 Main Protease Inhibitors: Enzyme Inhibition, Structure-Activity Relationships, Antiviral Activity, and X-ray Structure Determination.
J.Med.Chem., 65, 2022
|
|
5SKX
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 NSP14 in complex with Z126932614 | | 分子名称: | 2-[(methylsulfonyl)methyl]-1H-benzimidazole, PHOSPHATE ION, Proofreading exoribonuclease nsp14, ... | | 著者 | Imprachim, N, Yosaatmadja, Y, von-Delft, F, Bountra, C, Gileadi, O, Newman, J.A. | | 登録日 | 2022-03-03 | | 公開日 | 2022-03-16 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (2.342 Å) | | 主引用文献 | PanDDA analysis group deposition
To Be Published
|
|
7CBT
 
 | | The crystal structure of SARS-CoV-2 main protease in complex with GC376 | | 分子名称: | (1S,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase | | 著者 | Shi, Y, Peng, G. | | 登録日 | 2020-06-13 | | 公開日 | 2020-09-02 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.346 Å) | | 主引用文献 | The preclinical inhibitor GS441524 in combination with GC376 efficaciously inhibited the proliferation of SARS-CoV-2 in the mouse respiratory tract.
Emerg Microbes Infect, 10, 2021
|
|
5RLU
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 helicase in complex with Z744754722 | | 分子名称: | 2-(thiophen-2-yl)-1H-imidazole, Helicase, PHOSPHATE ION, ... | | 著者 | Newman, J.A, Yosaatmadja, Y, Douangamath, A, Aimon, A, Powell, A.J, Dias, A, Fearon, D, Dunnett, L, Brandao-Neto, J, Krojer, T, Skyner, R, Gorrie-Stone, T, Thompson, W, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | 登録日 | 2020-09-16 | | 公開日 | 2020-09-30 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (2.347 Å) | | 主引用文献 | Structure, mechanism and crystallographic fragment screening of the SARS-CoV-2 NSP13 helicase.
Nat Commun, 12, 2021
|
|
7GN7
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with ALP-POS-1cbc2fae-2 (Mpro-P2381) | | 分子名称: | (4S)-6-chloro-4-ethyl-N-(isoquinolin-4-yl)-1,1-dioxo-1,2,3,4-tetrahydro-1lambda~6~,2-benzothiazine-4-carboxamide, 3C-like proteinase, CHLORIDE ION, ... | | 著者 | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | 登録日 | 2023-08-11 | | 公開日 | 2023-11-08 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (2.349 Å) | | 主引用文献 | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
7XQ7
 
 | | The complex structure of WT-Mpro | | 分子名称: | 3C-like proteinase nsp5, SODIUM ION | | 著者 | Sahoo, P, Lenka, D.R, Kumar, A. | | 登録日 | 2022-05-06 | | 公開日 | 2023-03-01 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | Detailed Insights into the Inhibitory Mechanism of New Ebselen Derivatives against Main Protease (M pro ) of Severe Acute Respiratory Syndrome Coronavirus-2 (SARS-CoV-2).
Acs Pharmacol Transl Sci, 6, 2023
|
|
8RV4
 
 | | SARS-CoV-2 nsp16-nsp10 in complex with SAM derivative inhibitor 2 | | 分子名称: | 2'-O-methyltransferase nsp16, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 5-[[(2~{S},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methylsulfanylmethyl]-2-phenyl-benzoic acid, ... | | 著者 | Kalnins, G. | | 登録日 | 2024-01-31 | | 公開日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | SARS-CoV-2 nsp16-nsp10 in complex with SAM derivative inhibitor 2
To Be Published
|
|
8RZC
 
 | | SARS-CoV-2 nsp16-nsp10 in complex with SAM derivative inhibitor 11 | | 分子名称: | 2'-O-methyltransferase nsp16, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 3-[[(2S,3S,4R,5R)-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methylsulfanylmethyl]-5-imidazol-1-yl-benzoic acid, ... | | 著者 | Kalnins, G. | | 登録日 | 2024-02-12 | | 公開日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | SARS-CoV-2 nsp16-nsp10 in complex with SAM derivative inhibitor 11
To Be Published
|
|
7C8U
 
 | | The crystal structure of COVID-19 main protease in complex with GC376 | | 分子名称: | (1S,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase | | 著者 | Luan, X, Shang, W, Wang, Y, Yin, W, Jiang, Y, Feng, S, Wang, Y, Liu, M, Zhou, R, Zhang, Z, Wang, F, Cheng, W, Gao, M, Wang, H, Wu, W, Tian, R, Tian, Z, Jin, Y, Jiang, H.W, Zhang, L, Xu, H.E, Zhang, S. | | 登録日 | 2020-06-03 | | 公開日 | 2020-06-24 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | The crystal structure of COVID-19 main protease in complex with GC376
To Be Published
|
|
6XDH
 
 | |
8BS2
 
 | | Room-temperature structure of SARS-CoV-2 Main protease at 104 MPa helium gas pressure in a sapphire capillary | | 分子名称: | 3C-like proteinase, DIMETHYL SULFOXIDE | | 著者 | Lieske, J, Saouane, S, Guenther, S, Reinke, P.Y.A, Rahmani Mashhour, A, Meents, A. | | 登録日 | 2022-11-24 | | 公開日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | High-pressure macromolecular crystallography to explore the conformational space of proteins
To Be Published
|
|
7DPV
 
 | | SARS-CoV-2 3CL protease (3CLpro) in complex with 7-O-methyl-dihydromyricetin | | 分子名称: | (2S,3S)-3,5-dihydroxy-7-methoxy-2-(3,4,5-trihydroxyphenyl)chroman-4-one, 3C-like proteinase | | 著者 | Su, H.X, Zhao, W.F, Xie, H, Li, M.J, Xu, Y.C. | | 登録日 | 2020-12-21 | | 公開日 | 2021-05-12 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | Identification of pyrogallol as a warhead in design of covalent inhibitors for the SARS-CoV-2 3CL protease.
Nat Commun, 12, 2021
|
|
7RNW
 
 | |
7T70
 
 | |
8BWU
 
 | | Crystal structure of SARS-CoV-2 nsp14 methyltransferase domain in complex with the SS148 inhibitor | | 分子名称: | (2~{S})-2-azanyl-4-[[(2~{S},3~{S},4~{R},5~{R})-5-(4-azanyl-5-cyano-pyrrolo[2,3-d]pyrimidin-7-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methylsulfanyl]butanoic acid, Transcription factor ETV6,Proofreading exoribonuclease nsp14, ZINC ION | | 著者 | Konkolova, E, Klima, M, Boura, E, Jin, J, Kaniskan, H.U, Han, Y, Vedadi, M. | | 登録日 | 2022-12-07 | | 公開日 | 2023-10-11 | | 最終更新日 | 2024-06-26 | | 実験手法 | X-RAY DIFFRACTION (2.36 Å) | | 主引用文献 | Application of established computational techniques to identify potential SARS-CoV-2 Nsp14-MTase inhibitors in low data regimes
Digit Discov, 2024
|
|
5SL5
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 NSP14 in complex with Z32014663 | | 分子名称: | N,N,2,3-tetramethylbenzamide, PHOSPHATE ION, Proofreading exoribonuclease nsp14, ... | | 著者 | Imprachim, N, Yosaatmadja, Y, von-Delft, F, Bountra, C, Gileadi, O, Newman, J.A. | | 登録日 | 2022-03-03 | | 公開日 | 2022-03-16 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (2.36 Å) | | 主引用文献 | PanDDA analysis group deposition
To Be Published
|
|
8DZ6
 
 | | Crystal Structure of SARS-CoV-2 Main protease mutant Q189K in complex with Nirmatrelvir | | 分子名称: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5, DIMETHYL SULFOXIDE | | 著者 | Noske, G.D, Oliva, G, Godoy, A.S. | | 登録日 | 2022-08-06 | | 公開日 | 2022-10-05 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.366 Å) | | 主引用文献 | Structural basis of nirmatrelvir and ensitrelvir activity against naturally occurring polymorphisms of the SARS-CoV-2 main protease.
J.Biol.Chem., 299, 2023
|
|
7BQ7
 
 | | Crystal structure of 2019-nCoV nsp16-nsp10 complex | | 分子名称: | 2'-O-methyltransferase, Non-structural protein 10, S-ADENOSYLMETHIONINE, ... | | 著者 | Yan, L.M, Huang, Y.C, Lou, Z.Y, Rao, Z.H. | | 登録日 | 2020-03-24 | | 公開日 | 2020-06-17 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.37 Å) | | 主引用文献 | Crystal structure of 2019-nCoV nsp16-nsp10 complex
To Be Published
|
|
